Genome-wide and panel-based cell-free DNA characterization of patients with resectable esophageal adenocarcinoma
Tom van den Ende, Ymke van der Pol, Aafke Creemers, Norbert Moldovan, Dries Boers, Mark I van Berge Henegouwen, Maarten CCM Hulshof, Saskia AGM Cillessen, Nicole CT van Grieken, D Michiel Pegtel, Sarah Derks, Maarten F Bijlsma, Florent Mouliere, Hanneke WM van Laarhoven
下载PDF
{"title":"Genome-wide and panel-based cell-free DNA characterization of patients with resectable esophageal adenocarcinoma","authors":"Tom van den Ende, Ymke van der Pol, Aafke Creemers, Norbert Moldovan, Dries Boers, Mark I van Berge Henegouwen, Maarten CCM Hulshof, Saskia AGM Cillessen, Nicole CT van Grieken, D Michiel Pegtel, Sarah Derks, Maarten F Bijlsma, Florent Mouliere, Hanneke WM van Laarhoven","doi":"10.1002/path.6175","DOIUrl":null,"url":null,"abstract":"<p>Circulating tumor DNA (ctDNA) holds promise in resectable esophageal adenocarcinoma (EAC) to predict patient outcome but is not yet sensitive enough to be clinically applicable. Our aim was to combine ctDNA mutation data with shallow whole-genome sequencing (sWGS)-derived copy number tumor fraction estimates (ichorCNA) to improve pathological response and survival prediction in EAC. In total, 111 stage II/III EAC patients with baseline (<i>n</i> = 111), post-neoadjuvant chemoradiotherapy (nCRT) (<i>n</i> = 68), and pre-surgery (<i>n</i> = 92) plasma samples were used for ctDNA characterization. sWGS (<5× coverage) was performed on all time-point samples, and copy number aberrations were estimated using ichorCNA. Baseline and pre-surgery samples were sequenced using a custom amplicon panel for mutation detection. Detection of baseline ctDNA was successful in 44.3% of patients by amplicon sequencing and 10.5% by ichorCNA. Combining both, ctDNA could be detected in 50.5% of patients. Baseline ctDNA positivity was related to higher T stage (cT3, 4) (<i>p</i> = 0.017). There was no relationship between pathological response and baseline ctDNA positivity. However, baseline ctDNA metrics (variant allele frequency > 1% or ichorCNA > 3%) were associated with a high risk of disease progression [HR = 2.23 (95% CI 1.22–4.07), <i>p</i> = 0.007]. The non-clearance of a baseline variant or ichorCNA > 3% in pre-surgery samples was related to early progression [HR = 4.58 (95% CI 2.22–9.46), <i>p</i> < 0.001]. Multi-signal analysis improves detection of ctDNA and can be used for prognostication of resectable EAC patients. Future studies should explore the potential of multi-modality sequencing for risk stratification and treatment adaptation based on ctDNA results. © 2023 The Authors. <i>The Journal of Pathology</i> published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.</p>","PeriodicalId":232,"journal":{"name":"The Journal of Pathology","volume":"261 3","pages":"286-297"},"PeriodicalIF":5.2000,"publicationDate":"2023-08-24","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/path.6175","citationCount":"0","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"The Journal of Pathology","FirstCategoryId":"3","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/path.6175","RegionNum":2,"RegionCategory":"医学","ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"ONCOLOGY","Score":null,"Total":0}
引用次数: 0
引用
批量引用
Abstract
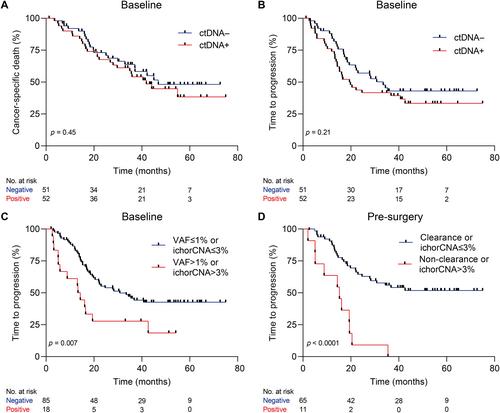
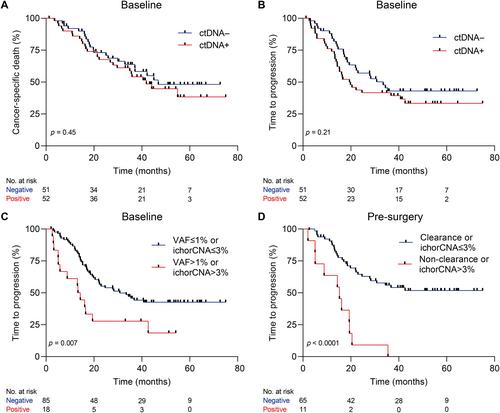
Circulating tumor DNA (ctDNA) holds promise in resectable esophageal adenocarcinoma (EAC) to predict patient outcome but is not yet sensitive enough to be clinically applicable. Our aim was to combine ctDNA mutation data with shallow whole-genome sequencing (sWGS)-derived copy number tumor fraction estimates (ichorCNA) to improve pathological response and survival prediction in EAC. In total, 111 stage II/III EAC patients with baseline (n = 111), post-neoadjuvant chemoradiotherapy (nCRT) (n = 68), and pre-surgery (n = 92) plasma samples were used for ctDNA characterization. sWGS (<5× coverage) was performed on all time-point samples, and copy number aberrations were estimated using ichorCNA. Baseline and pre-surgery samples were sequenced using a custom amplicon panel for mutation detection. Detection of baseline ctDNA was successful in 44.3% of patients by amplicon sequencing and 10.5% by ichorCNA. Combining both, ctDNA could be detected in 50.5% of patients. Baseline ctDNA positivity was related to higher T stage (cT3, 4) (p = 0.017). There was no relationship between pathological response and baseline ctDNA positivity. However, baseline ctDNA metrics (variant allele frequency > 1% or ichorCNA > 3%) were associated with a high risk of disease progression [HR = 2.23 (95% CI 1.22–4.07), p = 0.007]. The non-clearance of a baseline variant or ichorCNA > 3% in pre-surgery samples was related to early progression [HR = 4.58 (95% CI 2.22–9.46), p < 0.001]. Multi-signal analysis improves detection of ctDNA and can be used for prognostication of resectable EAC patients. Future studies should explore the potential of multi-modality sequencing for risk stratification and treatment adaptation based on ctDNA results. © 2023 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
可切除食管腺癌患者的全基因组和基于面板的无细胞DNA特征
循环肿瘤DNA(ctDNA)在可切除食管腺癌(EAC)中有望预测患者的预后,但其敏感性尚不足以应用于临床。我们的目的是将ctDNA突变数据与浅层全基因组测序(sWGS)衍生的拷贝数肿瘤分数估计(ichorCNA)相结合,以改善EAC的病理反应和生存预测。总共有111名Ⅱ/Ⅲ期EAC患者基线(n = 111)、新辅助放化疗后(nCRT)(n = 68)和手术前(n = 92)血浆样品用于ctDNA表征。对所有时间点样本进行sWGS(<;5×覆盖),并使用ichorCNA估计拷贝数像差。使用定制扩增子面板对基线和手术前样本进行测序,用于突变检测。44.3%的患者通过扩增子测序成功检测到基线ctDNA,10.5%的患者通过ichorCNA成功检测到。将两者结合,可在50.5%的患者中检测到ctDNA。基线ctDNA阳性与较高的T分期(cT3,4)有关(p = 0.017)。病理反应与基线ctDNA阳性之间没有关系。然而,基线ctDNA指标(变异等位基因频率 >; 1%或ichorCNA >; 3%)与疾病进展的高风险相关[HR = 2.23(95%置信区间1.22–4.07),p = 0.007]。基线变体或ichorCNA的未清除 >; 术前样本中3%与早期进展有关[HR = 4.58(95%置信区间2.22–9.46),p <; 0.001]。多信号分析提高了ctDNA的检测,可用于可切除EAC患者的预后。未来的研究应基于ctDNA结果探索多模式测序在风险分层和治疗适应方面的潜力。©2023作者。病理学杂志由John Wiley&;代表大不列颠及爱尔兰病理学会的Sons有限公司。
本文章由计算机程序翻译,如有差异,请以英文原文为准。