Esophageal squamous cell carcinoma (ESCC) has poor survival. Effective prognostic models with high application value remain lack.
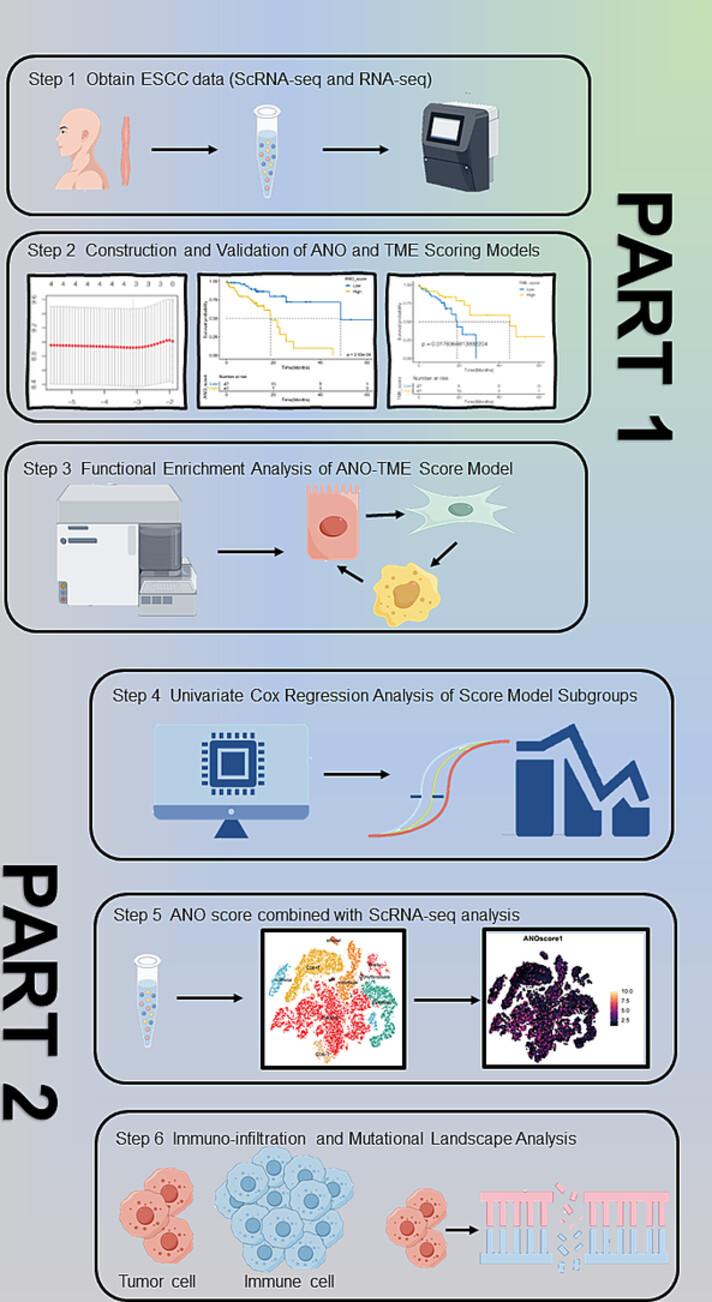
Bulk RNA seq and single cell RNA-seq data were retrieved from the XENA-TCGA-ESCC cohort and GSE188900. The anoikis-related gene score (ANO score) model and tumor microenvironment score (TME score) model were constructed and merged into three subgroups. Functional annotation was analyzed by Gene Ontology terms. Univariate and multivariate Cox regression analysis, least absolute shrinkage and selection operator regression analysis and weighted gene coexpression network analysis were performed to construct prognostic prediction models and identify prognostic value. Kaplan–Meier survival curves were drawn for evaluating the overall survival (OS) of patients classified by different score subgroups. Immunotherapy response and mutation analyses were also conducted.
In the ANO score model, TNFSF10 was an independent factor for the prognosis of ESCC patients. The area under the curve values of the ANO–TME score model in predicting the OS were 0.638 at 5 years and 0.632 at 7 years. Patients in the ANO low score–TME high score group had a much longer OS than patients in any other ANO–TME score subgroup (p < 0.001), suggesting a higher prognostic value. The differentially expressed genes of the ANO low score–TME high score group were mainly involved in cell adhesion molecules, nucleotide excision repair, the TGF-β signaling pathway and mismatch repair. TP53 (92%), TTN (38%) and NFE2L2 (31%) were the top genes with highest mutant frequency in the ANO low score–TME high score group.
A novel prognostic prediction model with high application value was constructed and identified for ESCC patients, which may provide evidence for immunotherapy in the treatment of ESCC.