Allergic diseases typically refer to a heterogeneous group of conditions primarily caused by the activation of mast cells or eosinophils, including atopic dermatitis (AD), allergic rhinitis (AR), and asthma. Asthma, AR, and AD collectively affect approximately one-fifth of the global population, imposing a significant economic burden on society. Despite the availability of drugs to treat allergic diseases, they have been shown to be insufficient in controlling relapses and halting disease progression. Therefore, new drug targets are needed to prevent the onset of allergic diseases.
We employed a Mendelian randomization approach to identify potential drug targets for the treatment of allergic diseases. Leveraging 1798 genetic instruments for 1537 plasma proteins from the latest reported Genome-Wide Association Studies (GWAS), we analyzed the GWAS summary statistics of Ferreira MA et al. (nCase = 180,129, nControl = 180,709) using the Mendelian randomization method. Furthermore, we validated our findings in the GWAS data from the FinnGen and UK Biobank cohorts. Subsequently, we conducted sensitivity tests through reverse causal analysis, Bayesian colocalization analysis, and phenotype scanning. Additionally, we performed protein-protein interaction analysis to determine the interaction between causal proteins. Finally, based on the potential protein targets, we conducted molecular docking to identify potential drugs for the treatment of allergic diseases.
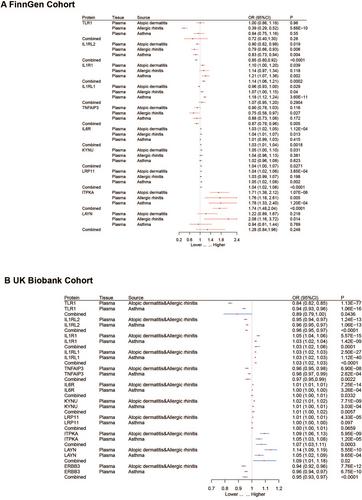
At Bonferroni significance (p < 3.25 × 10−5), the Mendelian randomization analysis revealed 11 significantly associated protein-allergic disease pairs. Among these, the increased levels of TNFAIP3, ERBB3, TLR1, and IL1RL2 proteins were associated with a reduced risk of allergic diseases, with corresponding odds ratios of 0.82 (0.76–0.88), 0.74 (0.66–0.82), 0.49 (0.45–0.55), and 0.81 (0.75–0.87), respectively. Conversely, increased levels of IL6R, IL1R1, ITPKA, IL1RL1, KYNU, LAYN, and LRP11 proteins were linked to an elevated risk of allergic diseases, with corresponding odds ratios of 1.04 (1.03–1.05), 1.25 (1.18–1.34), 1.48 (1.25–1.75), 1.14 (1.11–1.18), 1.09 (1.05–1.12), 1.96 (1.56–2.47), and 1.05 (1.03–1.07), respectively. Bayesian colocalization analysis suggested that LAYN (coloc.abf-PPH4 = 0.819) and TNFAIP3 (coloc.abf-PPH4 = 0.930) share the same variant associated with allergic diseases.
Our study demonstrates a causal association between the expression levels of TNFAIP3 and LAYN and the risk of allergic diseases, suggesting them as potential drug targets for these conditions, warranting further clinical investigation.