Previous studies have hinted at a significant link between lung cancer and the gut microbiome, yet their causal relationship remains to be elucidated.
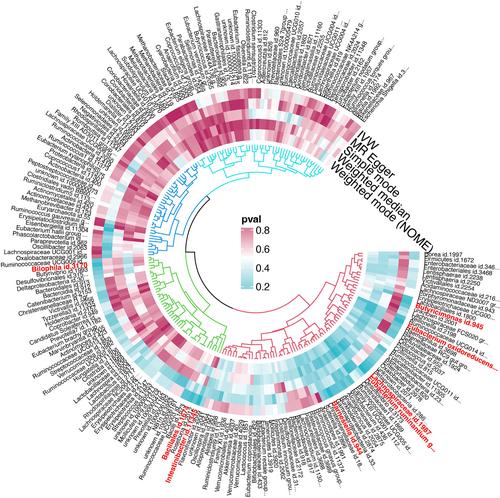
GWAS data for small cell lung cancer (SCLC) was extracted from the FinnGen consortium, comprising 179 cases and 218 613 controls. Genetic variation data for 211 gut microbiota were obtained as instrumental variables from MiBioGen. Mendelian randomization (MR) was employed to determine the causal relationship between the two, with inverse variance weighting (IVW) being the primary method for causal analysis. The MR results were validated through several sensitivity analyses.
The study identified a protective effect against SCLC for the genus Eubacterium ruminantium group (OR = 0.413, 95% CI: 0.223–0.767, p = 0.00513), genus Barnesiella (OR = 0.208, 95% CI: 0.0640–0.678, p = 0.00919), family Lachnospiraceae (OR = 0.319, 95% CI: 0.107–0.948, p = 0.03979), and genus Butyricimonas (OR = 0.376, 95% CI: 0.144–0.984, p = 0.04634). Conversely, genus Intestinibacter (OR = 3.214, 95% CI: 1.303–7.926, p = 0.01125), genus Eubacterium oxidoreducens group (OR = 3.391, 95% CI: 1.215–9.467, p = 0.01973), genus Bilophila (OR = 3.547, 95% CI: 1.106–11.371, p = 0.03315), and order Bacillales (OR = 1.860, 95% CI: 1.034–3.347, p = 0.03842) were found to potentially promote the onset of SCLC.
We identified potential causal relationships between certain gut microbiota and SCLC, offering new insights into microbiome-mediated mechanisms of SCLC pathogenesis, resistance, mutations, and more.