{"title":"Establishment of Transgene-Free Porcine Induced Pluripotent Stem Cells","authors":"J. Vanessa Conrad, Jaime A. Neira, Margaret Rusteika, Susanne Meyer, Dennis O. Clegg, Li-Fang Chu","doi":"10.1002/cpz1.1012","DOIUrl":null,"url":null,"abstract":"<p>Although protocols to generate authentic transgene-free mouse and human induced pluripotent stem cells (iPSCs) are now well established, standard methods for reprogramming porcine somatic cells still suffer from low efficiency and transgene retention. The Basic Protocol describes reprogramming procedures to establish transgene-free porcine iPSCs (PiPSCs) from porcine fibroblasts. This method uses episomal plasmids encoding <i>POU5F1</i>, <i>SOX2</i>, <i>NANOG</i>, <i>KLF4</i>, <i>SV40LT</i>, <i>c-MYC</i>, <i>LIN28A</i>, and <i>microRNA-302/367</i>, combined with an optimized medium, to establish PiPSC lines. Support protocols describe the establishment and characterization of clonal PiPSC lines, as well as the preparation of feeder cells and <i>EBNA1</i> mRNA. This optimized, step-by-step approach tailored to this species enables the efficient derivation of PiPSCs in ∼4 weeks. The establishment of transgene-free PiPSCs provides a new and valuable model for studies of larger mammalian species’ development, disease, and regenerative biology. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol</b>: Reprogramming of porcine fibroblasts with episomal plasmids</p><p><b>Support Protocol 1</b>: Preparation of mouse embryonic fibroblasts for feeder layer</p><p><b>Support Protocol 2</b>: Preparation of <i>in vitro</i>–transcribed <i>EBNA1</i> mRNA</p><p><b>Support Protocol 3</b>: Establishment of clonal porcine induced pluripotent stem cell (PiPSC) lines</p><p><b>Support Protocol 4</b>: PiPSC characterization: Genomic DNA PCR and RT-PCR</p><p><b>Support Protocol 5</b>: PiPSC characterization: Immunostaining</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 5","pages":""},"PeriodicalIF":2.2000,"publicationDate":"2024-05-07","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.1012","citationCount":"0","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/cpz1.1012","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
Abstract
Although protocols to generate authentic transgene-free mouse and human induced pluripotent stem cells (iPSCs) are now well established, standard methods for reprogramming porcine somatic cells still suffer from low efficiency and transgene retention. The Basic Protocol describes reprogramming procedures to establish transgene-free porcine iPSCs (PiPSCs) from porcine fibroblasts. This method uses episomal plasmids encoding POU5F1, SOX2, NANOG, KLF4, SV40LT, c-MYC, LIN28A, and microRNA-302/367, combined with an optimized medium, to establish PiPSC lines. Support protocols describe the establishment and characterization of clonal PiPSC lines, as well as the preparation of feeder cells and EBNA1 mRNA. This optimized, step-by-step approach tailored to this species enables the efficient derivation of PiPSCs in ∼4 weeks. The establishment of transgene-free PiPSCs provides a new and valuable model for studies of larger mammalian species’ development, disease, and regenerative biology. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol: Reprogramming of porcine fibroblasts with episomal plasmids
Support Protocol 1: Preparation of mouse embryonic fibroblasts for feeder layer
Support Protocol 2: Preparation of in vitro–transcribed EBNA1 mRNA
Support Protocol 3: Establishment of clonal porcine induced pluripotent stem cell (PiPSC) lines
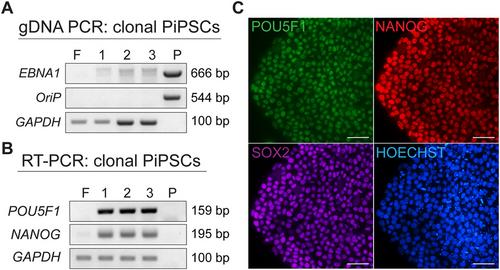
Support Protocol 4: PiPSC characterization: Genomic DNA PCR and RT-PCR
Support Protocol 5: PiPSC characterization: Immunostaining