Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His6-SUMO Tag Removal, Nucleotide Binding, and Crystal Growth
Li Lei, Joel M. Harp, John C. Chaput, Kelly Wassarman, Mark K. Schlegel, Muthiah Manoharan, Martin Egli
{"title":"Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His6-SUMO Tag Removal, Nucleotide Binding, and Crystal Growth","authors":"Li Lei, Joel M. Harp, John C. Chaput, Kelly Wassarman, Mark K. Schlegel, Muthiah Manoharan, Martin Egli","doi":"10.1002/cpz1.1088","DOIUrl":null,"url":null,"abstract":"<p>The middle (MID) domain of eukaryotic Argonaute (Ago) proteins and archaeal and bacterial homologues mediates the interaction with the 5′-terminal nucleotide of miRNA and siRNA guide strands. The MID domain of human Ago2 (hAgo2) is comprised of 139 amino acids with a molecular weight of 15.56 kDa. MID adopts a Rossman-like beta1-alpha1-beta2-alpha2-beta3-alpha3-beta4-alpha4 fold with a nucleotide specificity loop between beta3 and alpha3. Multiple crystal structures of nucleotides bound to hAgo2 MID have been reported, whereby complexes were obtained by soaking ligands into crystals of MID domain alone. This protocol describes a simplified one-step approach to grow well-diffracting crystals of hAgo2 MID-nucleotide complexes by mixing purified His<sub>6</sub>-SUMO-MID fusion protein, Ulp1 protease, and excess nucleotide in the presence of buffer and precipitant. The crystal structures of MID complexes with UMP, UTP and 2′-3′ linked α-<span>L</span>-threofuranosyl thymidine-3′-triphosphate (tTTP) are presented. This article also describes fluorescence-based assays to measure dissociation constants (<i>K<sub>d</sub></i>) of MID-nucleotide interactions for nucleoside 5′-monophosphates and nucleoside 3′,5′-bisphosphates. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Crystallization of Ago2 MID-nucleotide complexes</p><p><b>Basic Protocol 2</b>: Measurement of dissociation constant <i>K<sub>d</sub></i> between Ago2 MID and nucleotides</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":null,"pages":null},"PeriodicalIF":0.0000,"publicationDate":"2024-06-24","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.1088","citationCount":"0","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.1088","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
Abstract
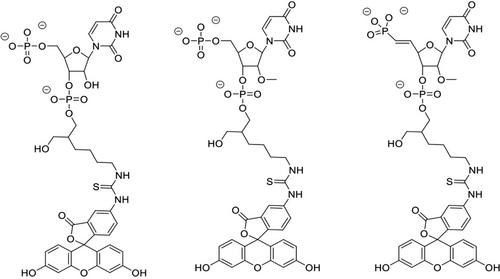
The middle (MID) domain of eukaryotic Argonaute (Ago) proteins and archaeal and bacterial homologues mediates the interaction with the 5′-terminal nucleotide of miRNA and siRNA guide strands. The MID domain of human Ago2 (hAgo2) is comprised of 139 amino acids with a molecular weight of 15.56 kDa. MID adopts a Rossman-like beta1-alpha1-beta2-alpha2-beta3-alpha3-beta4-alpha4 fold with a nucleotide specificity loop between beta3 and alpha3. Multiple crystal structures of nucleotides bound to hAgo2 MID have been reported, whereby complexes were obtained by soaking ligands into crystals of MID domain alone. This protocol describes a simplified one-step approach to grow well-diffracting crystals of hAgo2 MID-nucleotide complexes by mixing purified His6-SUMO-MID fusion protein, Ulp1 protease, and excess nucleotide in the presence of buffer and precipitant. The crystal structures of MID complexes with UMP, UTP and 2′-3′ linked α-L-threofuranosyl thymidine-3′-triphosphate (tTTP) are presented. This article also describes fluorescence-based assays to measure dissociation constants (Kd) of MID-nucleotide interactions for nucleoside 5′-monophosphates and nucleoside 3′,5′-bisphosphates. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1: Crystallization of Ago2 MID-nucleotide complexes
Basic Protocol 2: Measurement of dissociation constant Kd between Ago2 MID and nucleotides
Ago2 MID-核苷酸复合物的结构和稳定性:一体化(滴)His6-SUMO 标记去除、核苷酸结合和晶体生长。
真核生物 Argonaute(Ago)蛋白以及古细菌和细菌同源物的中间(MID)结构域介导与 miRNA 和 siRNA 引导链 5'-terminal 核苷酸的相互作用。人类 Ago2(hAgo2)的 MID 结构域由 139 个氨基酸组成,分子量为 15.56 kDa。MID 采用类似 Rossman 的 beta1-α1-beta2-α2-beta3-α3-beta4-α4 折叠结构,在 beta3 和 alpha3 之间有一个核苷酸特异性环。已报道了多种核苷酸与 hAgo2 MID 结合的晶体结构,其中的复合物是通过将配体浸泡到单独的 MID 结构域晶体中获得的。本方案描述了一种简化的一步法,通过将纯化的 His6-SUMO-MID 融合蛋白、Ulp1 蛋白酶和过量的核苷酸在缓冲液和沉淀剂的存在下混合,生长出衍射良好的 hAgo2 MID 核苷酸复合物晶体。文章展示了 MID 与 UMP、UTP 和 2'-3' 链接的 α-L- 苏呋糖基胸苷-3'-三磷酸(ttp)复合物的晶体结构。本文还介绍了基于荧光的检测方法,用于测量核苷-5'-单磷酸和核苷-3',5'-二磷酸的 MID-核苷酸相互作用的解离常数(Kd)。© 2024 作者。当前协议》由 Wiley Periodicals LLC 出版。基本方案 1:Ago2 MID-核苷酸复合物的结晶 基本方案 2:测量 Ago2 MID 与核苷酸之间的解离常数 Kd。
本文章由计算机程序翻译,如有差异,请以英文原文为准。


