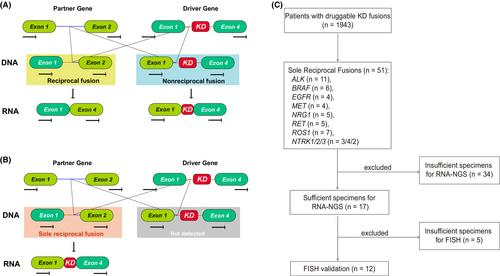
Building on our prior work that RNA alternative splicing modulates the druggability of kinase fusions, this study probes the clinical significance of sole reciprocal fusions. These rare genomic arrangements, despite lacking kinase domains at the DNA level, demonstrated potential RNA-level druggability in sporadic cases from our prior research.
Utilizing the large-scale multicenter approach, we performed RNA sequencing and clinical follow-up to evaluate a broad spectrum of kinase fusions, including ALK, ROS1, RET, BRAF, NTRK, MET, NRG1, and EGFR, in 1943 patients.
Our findings revealed 51 instances (2.57%) of sole reciprocal fusions, predominantly in lung (57%), colorectal (14%), and glioma (10%) cancers. Comparative analysis with an MSKCC cohort confirmed the prevalence in diverse cancer types and identified unique fusion partners and chromosomal locales. Cross-validation through RNA-NGS and FISH authenticated the existence of functional kinase domains in subsets including ALK, ROS1, RET, and BRAF, which correlated with positive clinical responses to targeted kinase inhibitors (KIs). Conversely, fusions involving EGFR, NRG1, and NTRK1/2/3 generated nonfunctional transcripts, suggesting the need for alternative therapeutic interventions.
This inaugural multicenter study introduces a novel algorithm for detecting and treating sole reciprocal fusions in advanced cancers, expanding the patient population potentially amenable to KIs.