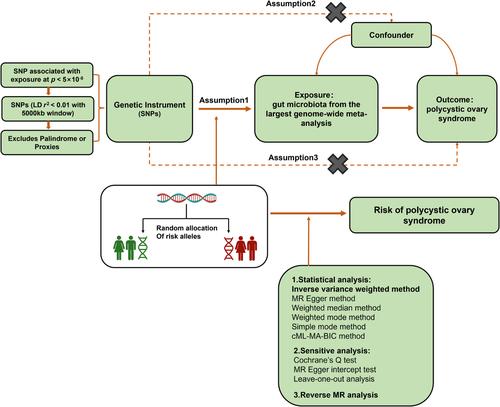
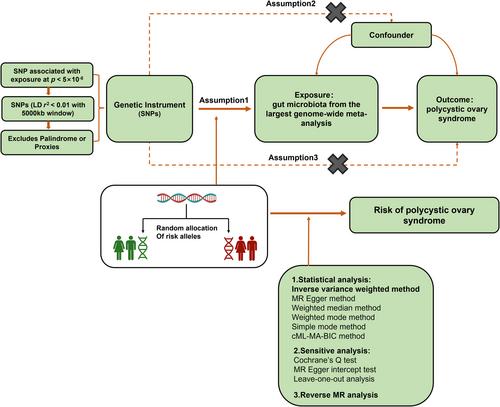
Previous studies have established a link between gut microbiota and polycystic ovary syndrome (PCOS), but little is known about their precise causal relationship. Therefore, this study aims to explore whether there are precise causal relationships between gut microbiota and PCOS.
We performed a bidirectional two-sample Mendelian randomization (MR) analysis. Datasets were from the largest published meta-analysis on gut microbiota composition and the FinnGen cohort of the IEU Open Genome-Wide Association Study Project database. Inverse variance weighted (IVW), MR-Egger, constrained maximum likelihood-based Mendelian randomization, weighted median, weighted mode, and simple mode were used. Cochran's Q and MR-Egger intercept tests were employed to measure the heterogeneity.
A total of 211 gut microbiota taxa were identified in MR analysis. Nine taxa of bacteria, including Alphaproteobacteria (0.55, 0.30–0.99, p = 0.04), Bacilli (1.76, 1.07–2.91, p = 0.03), Bilophila (0.42, 0.23–0.77, p < 0.01), Blautia (0.16, 0.03–0.79, p = 0.02), Burkholderiales (2.37, 1.22–4.62, p = 0.01), Candidatus Soleaferrea (0.65, 0.43–0.98, p = 0.04), Cyanobacteria (0.51, 0.31–0.83, p = 0.01), Holdemania (0.53, 0.35–0.81, p < 0.01), and Lachnospiraceae (1.86, 1.04–3.35, p = 0.03), were found to be associated with PCOS in the above MR methods included at least IVW method. Cochran's Q statistics and MR-Egger intercept test suggested no significant heterogeneity. In addition, 69 taxa were shown significant for at least the IVW method in reverse MR analysis, of these, 25 had a positive correlation, and 37 had a negative correlation. Additionally, Alphaproteobacteria and Lachnospiraceae (0.95, 0.91–0.98, p < 0.01; 0.97, 0.94–0.99, p = 0.02, respectively) were shown a bidirected causally association with PCOS.
Our study provides evidence of the bidirectional causal association between gut microbiota and PCOS from a genetic perspective.