{"title":"Generation of Barcode-Tagged Vibrio fischeri Deletion Strains and Barcode Sequencing (BarSeq) for Multiplex Strain Competitions","authors":"Hector L. Burgos, Mark J. Mandel","doi":"10.1002/cpz1.70024","DOIUrl":null,"url":null,"abstract":"<p><i>Vibrio fischeri</i> is a model mutualist for studying molecular processes affecting microbial colonization of animal hosts. We present a detailed protocol for a barcode sequencing (BarSeq) approach that combines targeted gene deletion with short-read sequencing technology to enable studies of mixed bacterial populations. This protocol includes wet lab steps to plan and produce the deletions, approaches to scale up mutant generation, protocols to prepare and conduct the strain competition, library preparation for sequencing on an Illumina iSeq 100 instrument, and data analysis with the barseq python package. Aspects of this protocol could be readily adapted for tagging wild-type <i>V. fischeri</i> strains with a neutral barcode for examination of population dynamics or BarSeq analyses in other species. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Production of the <i>erm-bar</i> DNA</p><p><b>Basic Protocol 2</b>: Generation of a targeted and barcoded deletion strain of <i>V. fischeri</i></p><p><b>Alternate Protocol</b>: Parallel generation of multiple barcode-tagged <i>V. fischeri</i> deletion strains</p><p><b>Basic Protocol 3</b>: Setting up mixed populations of barcode-tagged strains</p><p><b>Basic Protocol 4</b>: Performing a competitive growth assay</p><p><b>Basic Protocol 5</b>: Amplicon library preparation and equimolar pooling</p><p><b>Basic Protocol 6</b>: Sequencing on Illumina iSeq 100</p><p><b>Basic Protocol 7</b>: BarSeq data analysis</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 10","pages":""},"PeriodicalIF":2.2000,"publicationDate":"2024-10-26","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70024","citationCount":"0","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/cpz1.70024","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
Abstract
Vibrio fischeri is a model mutualist for studying molecular processes affecting microbial colonization of animal hosts. We present a detailed protocol for a barcode sequencing (BarSeq) approach that combines targeted gene deletion with short-read sequencing technology to enable studies of mixed bacterial populations. This protocol includes wet lab steps to plan and produce the deletions, approaches to scale up mutant generation, protocols to prepare and conduct the strain competition, library preparation for sequencing on an Illumina iSeq 100 instrument, and data analysis with the barseq python package. Aspects of this protocol could be readily adapted for tagging wild-type V. fischeri strains with a neutral barcode for examination of population dynamics or BarSeq analyses in other species. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
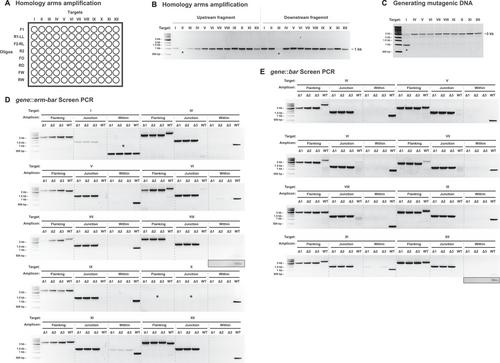
Basic Protocol 1: Production of the erm-bar DNA
Basic Protocol 2: Generation of a targeted and barcoded deletion strain of V. fischeri
Alternate Protocol: Parallel generation of multiple barcode-tagged V. fischeri deletion strains
Basic Protocol 3: Setting up mixed populations of barcode-tagged strains
Basic Protocol 4: Performing a competitive growth assay
Basic Protocol 5: Amplicon library preparation and equimolar pooling
Basic Protocol 6: Sequencing on Illumina iSeq 100
Basic Protocol 7: BarSeq data analysis

生成条形码标记的鱼腥弧菌缺失菌株和条形码测序(BarSeq),用于多重菌株竞赛。
费氏弧菌是研究影响微生物在动物宿主体内定殖的分子过程的互惠模式。我们介绍了条形码测序(BarSeq)方法的详细方案,该方法将靶向基因缺失与短读测序技术相结合,可用于混合细菌种群的研究。该方案包括计划和生产基因缺失的湿实验室步骤、扩大突变体生成规模的方法、准备和进行菌株竞争的方案、在 Illumina iSeq 100 仪器上进行测序的文库准备以及使用 barseq python 软件包进行数据分析。该方案的某些方面可随时用于用中性条形码标记野生型 V. fischeri 菌株,以检查其他物种的种群动态或进行 BarSeq 分析。© 2024 作者。当前协议》由 Wiley Periodicals LLC 出版。基本方案 1:生产 erm-bar DNA 基本方案 2:生成 V. fischeri 的目标和条形码缺失菌株 替代方案:基本方案 3:建立条形码标记菌株的混合种群 基本方案 4:进行竞争性生长试验 基本方案 5:扩增子文库制备和等摩尔汇集 基本方案 6:在 Illumina iSeq 100 上进行测序 基本方案 7:BarSeq 数据分析。
本文章由计算机程序翻译,如有差异,请以英文原文为准。



