QIIME 2 Enables Comprehensive End-to-End Analysis of Diverse Microbiome Data and Comparative Studies with Publicly Available Data
Mehrbod Estaki, Lingjing Jiang, Nicholas A. Bokulich, Daniel McDonald, Antonio González, Tomasz Kosciolek, Cameron Martino, Qiyun Zhu, Amanda Birmingham, Yoshiki Vázquez-Baeza, Matthew R. Dillon, Evan Bolyen, J. Gregory Caporaso, Rob Knight
下载PDF
{"title":"QIIME 2 Enables Comprehensive End-to-End Analysis of Diverse Microbiome Data and Comparative Studies with Publicly Available Data","authors":"Mehrbod Estaki, Lingjing Jiang, Nicholas A. Bokulich, Daniel McDonald, Antonio González, Tomasz Kosciolek, Cameron Martino, Qiyun Zhu, Amanda Birmingham, Yoshiki Vázquez-Baeza, Matthew R. Dillon, Evan Bolyen, J. Gregory Caporaso, Rob Knight","doi":"10.1002/cpbi.100","DOIUrl":null,"url":null,"abstract":"<p>QIIME 2 is a completely re-engineered microbiome bioinformatics platform based on the popular QIIME platform, which it has replaced. QIIME 2 facilitates comprehensive and fully reproducible microbiome data science, improving accessibility to diverse users by adding multiple user interfaces. QIIME 2 can be combined with Qiita, an open-source web-based platform, to re-use available data for meta-analysis. The following basic protocol describes how to install QIIME 2 on a single computer and analyze microbiome sequence data, from processing of raw DNA sequence reads through generating publishable interactive figures. These interactive figures allow readers of a study to interact with data with the same ease as its authors, advancing microbiome science transparency and reproducibility. We also show how plug-ins developed by the community to add analysis capabilities can be installed and used with QIIME 2, enhancing various aspects of microbiome analyses—e.g., improving taxonomic classification accuracy. Finally, we illustrate how users can perform meta-analyses combining different datasets using readily available public data through Qiita. In this tutorial, we analyze a subset of the Early Childhood Antibiotics and the Microbiome (ECAM) study, which tracked the microbiome composition and development of 43 infants in the United States from birth to 2 years of age, identifying microbiome associations with antibiotic exposure, delivery mode, and diet. For more information about QIIME 2, see https://qiime2.org. To troubleshoot or ask questions about QIIME 2 and microbiome analysis, join the active community at https://forum.qiime2.org. © 2020 The Authors.</p><p><b>Basic Protocol</b>: Using QIIME 2 with microbiome data</p><p><b>Support Protocol</b>: Further microbiome analyses</p>","PeriodicalId":10958,"journal":{"name":"Current protocols in bioinformatics","volume":"70 1","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2020-04-28","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cpbi.100","citationCount":"165","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols in bioinformatics","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpbi.100","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"Biochemistry, Genetics and Molecular Biology","Score":null,"Total":0}
引用次数: 165
引用
批量引用
Abstract
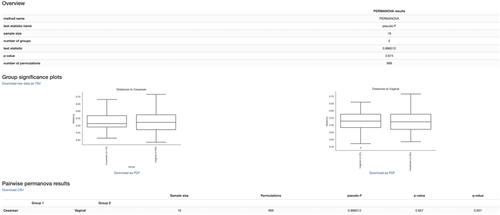
QIIME 2 is a completely re-engineered microbiome bioinformatics platform based on the popular QIIME platform, which it has replaced. QIIME 2 facilitates comprehensive and fully reproducible microbiome data science, improving accessibility to diverse users by adding multiple user interfaces. QIIME 2 can be combined with Qiita, an open-source web-based platform, to re-use available data for meta-analysis. The following basic protocol describes how to install QIIME 2 on a single computer and analyze microbiome sequence data, from processing of raw DNA sequence reads through generating publishable interactive figures. These interactive figures allow readers of a study to interact with data with the same ease as its authors, advancing microbiome science transparency and reproducibility. We also show how plug-ins developed by the community to add analysis capabilities can be installed and used with QIIME 2, enhancing various aspects of microbiome analyses—e.g., improving taxonomic classification accuracy. Finally, we illustrate how users can perform meta-analyses combining different datasets using readily available public data through Qiita. In this tutorial, we analyze a subset of the Early Childhood Antibiotics and the Microbiome (ECAM) study, which tracked the microbiome composition and development of 43 infants in the United States from birth to 2 years of age, identifying microbiome associations with antibiotic exposure, delivery mode, and diet. For more information about QIIME 2, see https://qiime2.org. To troubleshoot or ask questions about QIIME 2 and microbiome analysis, join the active community at https://forum.qiime2.org. © 2020 The Authors.
Basic Protocol : Using QIIME 2 with microbiome data
Support Protocol : Further microbiome analyses