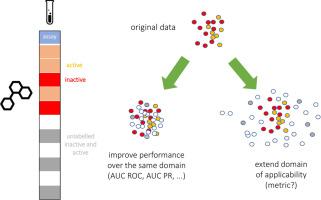
In a drug discovery setting, pharmaceutical companies own substantial but confidential datasets. The MELLODDY project developed a privacy-preserving federated machine learning solution and deployed it at an unprecedented scale. Each partner built models for their own private assays that benefitted from a shared representation. Established predictive performance metrics such as AUC ROC or AUC PR are constrained to unseen labeled chemical space and cannot gage performance gains in unlabeled chemical space. Federated learning indirectly extends labeled space, but in a privacy-preserving context, a partner cannot use this label extension for performance assessment. Metrics that estimate uncertainty on a prediction can be calculated even where no label is known. Practically, the chemical space covered with predictions above an uncertainty threshold, reflects the applicability domain of a model. After establishing a link to established performance metrics, we propose the efficiency from the conformal prediction framework (‘conformal efficiency’) as a proxy to the applicability domain size. A documented extension of the applicability domain would qualify as a tangible benefit from federated learning. In interim assessments, MELLODDY partners reported a median increase in conformal efficiency of the federated over the single-partner model of 5.5% (with increases up to 9.7%). Subject to distributional conditions, that efficiency increase can be directly interpreted as the expected increase in conformal i.e. low uncertainty predictions. In conclusion, we present the first indication that privacy-preserving federated machine learning across massive drug-discovery datasets from ten pharma partners indeed extends the applicability domain of property prediction models.