下载PDF
{"title":"PROMIS: Global Analysis of PROtein-Metabolite Interactions","authors":"Ewelina M. Sokolowska, Dennis Schlossarek, Marcin Luzarowski, Aleksandra Skirycz","doi":"10.1002/cppb.20101","DOIUrl":null,"url":null,"abstract":"<p>Small molecules are not only intermediates of metabolism, but also play important roles in signaling and in controlling cellular metabolism, growth, and development. Although a few systematic studies have been conducted, the true extent of protein–small molecule interactions in biological systems remains unknown. PROtein–metabolite interactions using size separation (PROMIS) is a method for studying protein–small molecule interactions in a non-targeted, proteome- and metabolome-wide manner. This approach uses size-exclusion chromatography followed by proteomics and metabolomics liquid chromatography–mass spectrometry analysis of the collected fractions. Assuming that small molecules bound to proteins would co-fractionate together, we found numerous small molecules co-eluting with proteins, strongly suggesting the formation of stable complexes. Using PROMIS, we identified known small molecule–protein complexes, such as between enzymes and cofactors, and also found novel interactions. © 2019 The Authors.</p><p><b>Basic Protocol 1</b>: Preparation of native cell lysate from plant material</p><p><b>Support Protocol</b>: Bradford assay to determine protein concentration</p><p><b>Basic Protocol 2</b>: Separation of molecular complexes using size-exclusion chromatography</p><p><b>Basic Protocol 3</b>: Simultaneous extraction of proteins and metabolites using single-step extraction protocol</p><p><b>Basic Protocol 4</b>: Metabolomics analysis</p><p><b>Basic Protocol 5</b>: Proteomics analysis</p>","PeriodicalId":10932,"journal":{"name":"Current protocols in plant biology","volume":"4 4","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2019-11-21","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cppb.20101","citationCount":"13","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols in plant biology","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cppb.20101","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"Agricultural and Biological Sciences","Score":null,"Total":0}
引用次数: 13
引用
批量引用
Abstract
Small molecules are not only intermediates of metabolism, but also play important roles in signaling and in controlling cellular metabolism, growth, and development. Although a few systematic studies have been conducted, the true extent of protein–small molecule interactions in biological systems remains unknown. PROtein–metabolite interactions using size separation (PROMIS) is a method for studying protein–small molecule interactions in a non-targeted, proteome- and metabolome-wide manner. This approach uses size-exclusion chromatography followed by proteomics and metabolomics liquid chromatography–mass spectrometry analysis of the collected fractions. Assuming that small molecules bound to proteins would co-fractionate together, we found numerous small molecules co-eluting with proteins, strongly suggesting the formation of stable complexes. Using PROMIS, we identified known small molecule–protein complexes, such as between enzymes and cofactors, and also found novel interactions. © 2019 The Authors.
Basic Protocol 1 : Preparation of native cell lysate from plant material
Support Protocol : Bradford assay to determine protein concentration
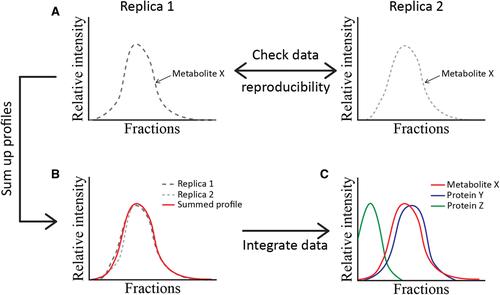
Basic Protocol 2 : Separation of molecular complexes using size-exclusion chromatography
Basic Protocol 3 : Simultaneous extraction of proteins and metabolites using single-step extraction protocol
Basic Protocol 4 : Metabolomics analysis
Basic Protocol 5 : Proteomics analysis
前景:蛋白质-代谢物相互作用的全局分析
小分子不仅是代谢的中间体,而且在信号传导和调控细胞代谢、生长发育等方面发挥着重要作用。尽管已经进行了一些系统的研究,但生物系统中蛋白质-小分子相互作用的真实程度仍然未知。利用大小分离技术(PROMIS)研究蛋白质-代谢物相互作用是一种非靶向、蛋白质组和代谢组范围内研究蛋白质-小分子相互作用的方法。该方法采用尺寸排除色谱法,然后使用蛋白质组学和代谢组学液相色谱-质谱法对收集的部分进行分析。假设与蛋白质结合的小分子会一起共分离,我们发现许多小分子与蛋白质共洗脱,这强烈表明形成了稳定的复合物。利用PROMIS,我们确定了已知的小分子-蛋白质复合物,例如酶和辅因子之间的复合物,并发现了新的相互作用。©2019作者。基本方案1:从植物材料中制备天然细胞裂解液支持方案:Bradford测定蛋白质浓度基本方案2:使用尺寸排除色谱分离分子复合物基本方案3:使用单步提取同时提取蛋白质和代谢物基本方案4:代谢组学分析基本方案5:蛋白质组学分析
本文章由计算机程序翻译,如有差异,请以英文原文为准。