Body composition is of clinical importance in colorectal cancer patients, but is rarely assessed because of time-consuming manual segmentation. We developed and tested BodySegAI, a deep learning-based software for automated body composition quantification from routinely acquired computed tomography (CT) scans.
A two-dimensional U-Net convolutional network was trained on 2989 abdominal CT slices from L2 to S1 to segment skeletal muscle (SM), visceral adipose tissue (VAT), subcutaneous adipose tissue (SAT), and intermuscular and intramuscular adipose tissue (IMAT). Human ground truth was established by combining segmentations from three human readers. BodySegAI was tested using 154 slices against the human ground truth and compared with a software named AutoMATiCA.
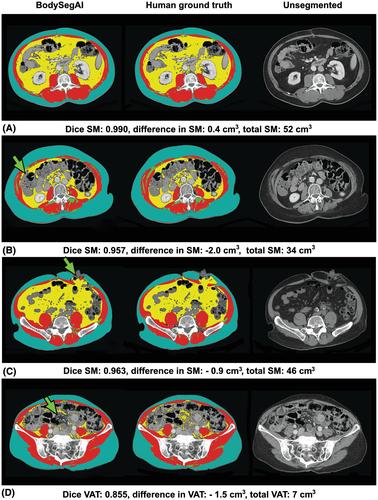
Median Dice scores for BodySegAI against human ground truth were 0.969, 0.814, 0.986, and 0.990 for SM, IMAT, VAT, and SAT, respectively. The mean differences per slice for SM were −0.09 cm3, IMAT: −0.17 cm3, VAT: −0.12 cm3, and SAT: 0.67 cm3. Median absolute errors for SM, IMAT, VAT, and SAT were 1.35, 10.54, 0.91, and 1.07%, respectively. When analysing different anatomical levels separately, L3 and S1 demonstrated the overall highest and lowest Dice scores, respectively. On average, BodySegAI segmented 148 times faster than human readers (4.9 vs. 726.5 seconds, P < 0.001). Also, BodySegAI presented higher Dice scores for SM, IMAT, SAT, and VAT than AutoMATiCA (slices = 154).
BodySegAI rapidly generates excellent segmentation of SM, VAT, and SAT and good segmentation of IMAT in L2 to S1 among colorectal cancer patients and may replace semi-manual segmentation.