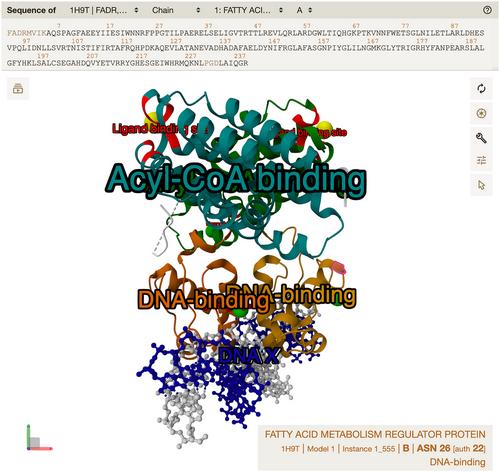
{"title":"使用 MolViewSpec 工具包描述和共享分子可视化。","authors":"Sebastian Bittrich, Adam Midlik, Mihaly Varadi, Sameer Velankar, Stephen K. Burley, Jasmine Y. Young, David Sehnal, Brinda Vallat","doi":"10.1002/cpz1.1099","DOIUrl":null,"url":null,"abstract":"<p>With the ever-expanding toolkit of molecular viewers, the ability to visualize macromolecular structures has never been more accessible. Yet, the idiosyncratic technical intricacies across tools and the integration complexities associated with handling structure annotation data present significant barriers to seamless interoperability and steep learning curves for many users. The necessity for reproducible data visualizations is at the forefront of the current challenges. Recently, we introduced MolViewSpec (homepage: https://molstar.org/mol-view-spec/, GitHub project: https://github.com/molstar/mol-view-spec), a specification approach that defines molecular visualizations, decoupling them from the varying implementation details of different molecular viewers. Through the protocols presented herein, we demonstrate how to use MolViewSpec and its 3D view–building Python library for creating sophisticated, customized 3D views covering all standard molecular visualizations. MolViewSpec supports representations like cartoon and ball-and-stick with coloring, labeling, and applying complex transformations such as superposition to any macromolecular structure file in mmCIF, BinaryCIF, and PDB formats. These examples showcase progress towards reusability and interoperability of molecular 3D visualization in an era when handling molecular structures at scale is a timely and pressing matter in structural bioinformatics as well as research and education across the life sciences. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Creating a MolViewSpec view using the MolViewSpec Python package</p><p><b>Basic Protocol 2</b>: Creating a MolViewSpec view with reference to MolViewSpec annotation files</p><p><b>Basic Protocol 3</b>: Creating a MolViewSpec view with labels and other advanced features</p><p><b>Support Protocol 1</b>: Computing rotation and translation vectors</p><p><b>Support Protocol 2</b>: Creating a MolViewSpec annotation file</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":null,"pages":null},"PeriodicalIF":0.0000,"publicationDate":"2024-07-18","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.1099","citationCount":"0","resultStr":"{\"title\":\"Describing and Sharing Molecular Visualizations Using the MolViewSpec Toolkit\",\"authors\":\"Sebastian Bittrich, Adam Midlik, Mihaly Varadi, Sameer Velankar, Stephen K. Burley, Jasmine Y. Young, David Sehnal, Brinda Vallat\",\"doi\":\"10.1002/cpz1.1099\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>With the ever-expanding toolkit of molecular viewers, the ability to visualize macromolecular structures has never been more accessible. Yet, the idiosyncratic technical intricacies across tools and the integration complexities associated with handling structure annotation data present significant barriers to seamless interoperability and steep learning curves for many users. The necessity for reproducible data visualizations is at the forefront of the current challenges. Recently, we introduced MolViewSpec (homepage: https://molstar.org/mol-view-spec/, GitHub project: https://github.com/molstar/mol-view-spec), a specification approach that defines molecular visualizations, decoupling them from the varying implementation details of different molecular viewers. Through the protocols presented herein, we demonstrate how to use MolViewSpec and its 3D view–building Python library for creating sophisticated, customized 3D views covering all standard molecular visualizations. MolViewSpec supports representations like cartoon and ball-and-stick with coloring, labeling, and applying complex transformations such as superposition to any macromolecular structure file in mmCIF, BinaryCIF, and PDB formats. These examples showcase progress towards reusability and interoperability of molecular 3D visualization in an era when handling molecular structures at scale is a timely and pressing matter in structural bioinformatics as well as research and education across the life sciences. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Creating a MolViewSpec view using the MolViewSpec Python package</p><p><b>Basic Protocol 2</b>: Creating a MolViewSpec view with reference to MolViewSpec annotation files</p><p><b>Basic Protocol 3</b>: Creating a MolViewSpec view with labels and other advanced features</p><p><b>Support Protocol 1</b>: Computing rotation and translation vectors</p><p><b>Support Protocol 2</b>: Creating a MolViewSpec annotation file</p>\",\"PeriodicalId\":93970,\"journal\":{\"name\":\"Current protocols\",\"volume\":null,\"pages\":null},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2024-07-18\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.1099\",\"citationCount\":\"0\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current protocols\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.1099\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"\",\"JCRName\":\"\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.1099","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0