{"title":"遗传操作光滑念珠菌。","authors":"Jane Usher","doi":"10.1002/cpz1.70014","DOIUrl":null,"url":null,"abstract":"<p><i>Candida glabrata</i> (<i>Nakaseomyces glabratus</i>) is an opportunistic fungal pathogen that has become a significant concern in clinical settings due to its increasing resistance to antifungal treatments. Understanding the genetic basis of its pathogenicity and resistance mechanisms is crucial for developing new therapeutic strategies. One powerful method of studying gene function is through targeted gene deletion. This paper outlines a comprehensive protocol for the deletion of genes in <i>C. glabrata</i>, encompassing primer design, preparation of electrocompetent cells, transformation, and finally confirmation of the gene deletion. The protocol begins with the identification and design of primers necessary for generating deletion constructs, involving the precise targeting of up- and downstream regions flanking the gene of interest to ensure high specificity and efficiency of homologous recombination. Followed is the preparation of electrocompetent cells, a critical step for successful transformation. Transformation of the competent cells is achieved through electroporation, facilitating the introduction of exogenous DNA into the cells. This is followed by the selection and confirmation of successfully transformed colonies. Confirmation involves the use of colony PCR to verify the correct integration of the NAT resistance cassette and deletion of the target gene. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Primer design for gene deletion in <i>C. glabrata</i></p><p><b>Basic Protocol 2</b>: Preparing competent <i>C. glabrata</i> cells</p><p><b>Basic Protocol 3</b>: Transforming <i>C. glabrata</i> using electroporation</p><p><b>Basic Protocol 4</b>: Confirming deletion strains with colony PCR</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 9","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2024-09-06","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70014","citationCount":"0","resultStr":"{\"title\":\"Genetic Manipulation of Candida glabrata\",\"authors\":\"Jane Usher\",\"doi\":\"10.1002/cpz1.70014\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p><i>Candida glabrata</i> (<i>Nakaseomyces glabratus</i>) is an opportunistic fungal pathogen that has become a significant concern in clinical settings due to its increasing resistance to antifungal treatments. Understanding the genetic basis of its pathogenicity and resistance mechanisms is crucial for developing new therapeutic strategies. One powerful method of studying gene function is through targeted gene deletion. This paper outlines a comprehensive protocol for the deletion of genes in <i>C. glabrata</i>, encompassing primer design, preparation of electrocompetent cells, transformation, and finally confirmation of the gene deletion. The protocol begins with the identification and design of primers necessary for generating deletion constructs, involving the precise targeting of up- and downstream regions flanking the gene of interest to ensure high specificity and efficiency of homologous recombination. Followed is the preparation of electrocompetent cells, a critical step for successful transformation. Transformation of the competent cells is achieved through electroporation, facilitating the introduction of exogenous DNA into the cells. This is followed by the selection and confirmation of successfully transformed colonies. Confirmation involves the use of colony PCR to verify the correct integration of the NAT resistance cassette and deletion of the target gene. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Primer design for gene deletion in <i>C. glabrata</i></p><p><b>Basic Protocol 2</b>: Preparing competent <i>C. glabrata</i> cells</p><p><b>Basic Protocol 3</b>: Transforming <i>C. glabrata</i> using electroporation</p><p><b>Basic Protocol 4</b>: Confirming deletion strains with colony PCR</p>\",\"PeriodicalId\":93970,\"journal\":{\"name\":\"Current protocols\",\"volume\":\"4 9\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2024-09-06\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70014\",\"citationCount\":\"0\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current protocols\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.70014\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"\",\"JCRName\":\"\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.70014","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
Genetic Manipulation of Candida glabrata
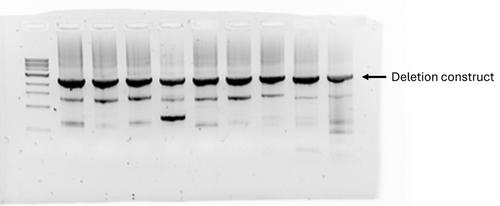
Candida glabrata (Nakaseomyces glabratus) is an opportunistic fungal pathogen that has become a significant concern in clinical settings due to its increasing resistance to antifungal treatments. Understanding the genetic basis of its pathogenicity and resistance mechanisms is crucial for developing new therapeutic strategies. One powerful method of studying gene function is through targeted gene deletion. This paper outlines a comprehensive protocol for the deletion of genes in C. glabrata, encompassing primer design, preparation of electrocompetent cells, transformation, and finally confirmation of the gene deletion. The protocol begins with the identification and design of primers necessary for generating deletion constructs, involving the precise targeting of up- and downstream regions flanking the gene of interest to ensure high specificity and efficiency of homologous recombination. Followed is the preparation of electrocompetent cells, a critical step for successful transformation. Transformation of the competent cells is achieved through electroporation, facilitating the introduction of exogenous DNA into the cells. This is followed by the selection and confirmation of successfully transformed colonies. Confirmation involves the use of colony PCR to verify the correct integration of the NAT resistance cassette and deletion of the target gene. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1: Primer design for gene deletion in C. glabrata
Basic Protocol 2: Preparing competent C. glabrata cells
Basic Protocol 3: Transforming C. glabrata using electroporation
Basic Protocol 4: Confirming deletion strains with colony PCR