下载PDF
{"title":"用IUPred2A分析蛋白质紊乱","authors":"Gábor Erdős, Zsuzsanna Dosztányi","doi":"10.1002/cpbi.99","DOIUrl":null,"url":null,"abstract":"<p>IUPred2A is a combined prediction tool designed to discover intrinsically disordered or conditionally disordered proteins and protein regions. Intrinsically disordered regions exist without a well-defined three-dimensional structure in isolation but carry out important biological functions. Over the years, various prediction methods have been developed to characterize disordered regions. The existence of disordered segments can also be dependent on different factors such as binding partners or environmental traits like pH or redox potential, and recognizing such regions represents additional computational challenges. In this article, we present detailed instructions on how to use IUPred2A, one of the most widely used tools for the prediction of disordered regions/proteins or conditionally disordered segments, and provide examples of how the predictions can be interpreted in different contexts. © 2020 The Authors.</p><p><b>Basic Protocol 1</b>: Analyzing disorder propensity with IUPred2A online</p><p><b>Basic Protocol 2</b>: Analyzing disordered binding regions using ANCHOR2</p><p><b>Support Protocol 1</b>: Interpretation of the results</p><p><b>Basic Protocol 3</b>: Analyzing redox-sensitive disordered regions</p><p><b>Support Protocol 2</b>: Download options</p><p><b>Support Protocol 3</b>: REST API for programmatic purposes</p><p><b>Basic Protocol 4</b>: Using IUPred2A locally</p>","PeriodicalId":10958,"journal":{"name":"Current protocols in bioinformatics","volume":"70 1","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2020-04-01","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cpbi.99","citationCount":"201","resultStr":"{\"title\":\"Analyzing Protein Disorder with IUPred2A\",\"authors\":\"Gábor Erdős, Zsuzsanna Dosztányi\",\"doi\":\"10.1002/cpbi.99\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>IUPred2A is a combined prediction tool designed to discover intrinsically disordered or conditionally disordered proteins and protein regions. Intrinsically disordered regions exist without a well-defined three-dimensional structure in isolation but carry out important biological functions. Over the years, various prediction methods have been developed to characterize disordered regions. The existence of disordered segments can also be dependent on different factors such as binding partners or environmental traits like pH or redox potential, and recognizing such regions represents additional computational challenges. In this article, we present detailed instructions on how to use IUPred2A, one of the most widely used tools for the prediction of disordered regions/proteins or conditionally disordered segments, and provide examples of how the predictions can be interpreted in different contexts. © 2020 The Authors.</p><p><b>Basic Protocol 1</b>: Analyzing disorder propensity with IUPred2A online</p><p><b>Basic Protocol 2</b>: Analyzing disordered binding regions using ANCHOR2</p><p><b>Support Protocol 1</b>: Interpretation of the results</p><p><b>Basic Protocol 3</b>: Analyzing redox-sensitive disordered regions</p><p><b>Support Protocol 2</b>: Download options</p><p><b>Support Protocol 3</b>: REST API for programmatic purposes</p><p><b>Basic Protocol 4</b>: Using IUPred2A locally</p>\",\"PeriodicalId\":10958,\"journal\":{\"name\":\"Current protocols in bioinformatics\",\"volume\":\"70 1\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2020-04-01\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://sci-hub-pdf.com/10.1002/cpbi.99\",\"citationCount\":\"201\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current protocols in bioinformatics\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cpbi.99\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"Q1\",\"JCRName\":\"Biochemistry, Genetics and Molecular Biology\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols in bioinformatics","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpbi.99","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"Biochemistry, Genetics and Molecular Biology","Score":null,"Total":0}
引用次数: 201
引用
批量引用
Analyzing Protein Disorder with IUPred2A
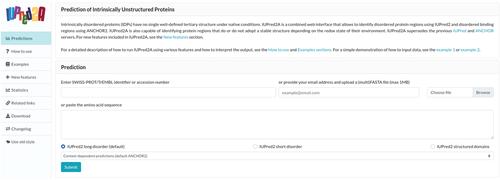
IUPred2A is a combined prediction tool designed to discover intrinsically disordered or conditionally disordered proteins and protein regions. Intrinsically disordered regions exist without a well-defined three-dimensional structure in isolation but carry out important biological functions. Over the years, various prediction methods have been developed to characterize disordered regions. The existence of disordered segments can also be dependent on different factors such as binding partners or environmental traits like pH or redox potential, and recognizing such regions represents additional computational challenges. In this article, we present detailed instructions on how to use IUPred2A, one of the most widely used tools for the prediction of disordered regions/proteins or conditionally disordered segments, and provide examples of how the predictions can be interpreted in different contexts. © 2020 The Authors.
Basic Protocol 1 : Analyzing disorder propensity with IUPred2A online
Basic Protocol 2 : Analyzing disordered binding regions using ANCHOR2
Support Protocol 1 : Interpretation of the results
Basic Protocol 3 : Analyzing redox-sensitive disordered regions
Support Protocol 2 : Download options
Support Protocol 3 : REST API for programmatic purposes
Basic Protocol 4 : Using IUPred2A locally