下载PDF
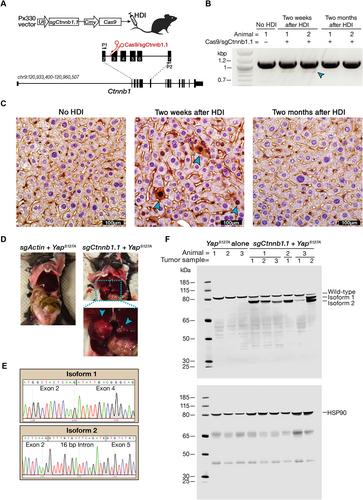
{"title":"crispr诱导的β-catenin外显子跳变揭示了驱动不同亚型肝癌的致瘤突变","authors":"Haiwei Mou, Onur Eskiocak, Kadir A. Özler, Megan Gorman, Junjiayu Yue, Ying Jin, Zhikai Wang, Ya Gao, Tobias Janowitz, Hannah V. Meyer, Tianxiong Yu, John E Wilkinson, Alper Kucukural, Deniz M. Ozata, Semir Beyaz","doi":"10.1002/path.6054","DOIUrl":null,"url":null,"abstract":"<p>CRISPR/Cas9-driven cancer modeling studies are based on the disruption of tumor suppressor genes by small insertions or deletions (indels) that lead to frame-shift mutations. In addition, CRISPR/Cas9 is widely used to define the significance of cancer oncogenes and genetic dependencies in loss-of-function studies. However, how CRISPR/Cas9 influences gain-of-function oncogenic mutations is elusive. Here, we demonstrate that single guide RNA targeting exon 3 of <i>Ctnnb1</i> (encoding β-catenin) results in exon skipping and generates gain-of-function isoforms <i>in vivo</i>. CRISPR/Cas9-mediated exon skipping of <i>Ctnnb1</i> induces liver tumor formation in synergy with YAP<sup>S127A</sup> in mice. We define two distinct exon skipping-induced tumor subtypes with different histological and transcriptional features. Notably, ectopic expression of two exon-skipped β-catenin transcript isoforms together with YAP<sup>S127A</sup> phenocopies the two distinct subtypes of liver cancer. Moreover, we identify similar <i>CTNNB1</i> exon-skipping events in patients with hepatocellular carcinoma. Collectively, our findings advance our understanding of β-catenin-related tumorigenesis and reveal that CRISPR/Cas9 can be repurposed, <i>in vivo</i>, to study gain-of-function mutations of oncogenes in cancer. © 2023 The Authors. <i>The Journal of Pathology</i> published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.</p>","PeriodicalId":232,"journal":{"name":"The Journal of Pathology","volume":"259 4","pages":"415-427"},"PeriodicalIF":5.6000,"publicationDate":"2023-01-15","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://pathsocjournals.onlinelibrary.wiley.com/doi/epdf/10.1002/path.6054","citationCount":"1","resultStr":"{\"title\":\"CRISPR-induced exon skipping of β-catenin reveals tumorigenic mutants driving distinct subtypes of liver cancer\",\"authors\":\"Haiwei Mou, Onur Eskiocak, Kadir A. Özler, Megan Gorman, Junjiayu Yue, Ying Jin, Zhikai Wang, Ya Gao, Tobias Janowitz, Hannah V. Meyer, Tianxiong Yu, John E Wilkinson, Alper Kucukural, Deniz M. Ozata, Semir Beyaz\",\"doi\":\"10.1002/path.6054\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>CRISPR/Cas9-driven cancer modeling studies are based on the disruption of tumor suppressor genes by small insertions or deletions (indels) that lead to frame-shift mutations. In addition, CRISPR/Cas9 is widely used to define the significance of cancer oncogenes and genetic dependencies in loss-of-function studies. However, how CRISPR/Cas9 influences gain-of-function oncogenic mutations is elusive. Here, we demonstrate that single guide RNA targeting exon 3 of <i>Ctnnb1</i> (encoding β-catenin) results in exon skipping and generates gain-of-function isoforms <i>in vivo</i>. CRISPR/Cas9-mediated exon skipping of <i>Ctnnb1</i> induces liver tumor formation in synergy with YAP<sup>S127A</sup> in mice. We define two distinct exon skipping-induced tumor subtypes with different histological and transcriptional features. Notably, ectopic expression of two exon-skipped β-catenin transcript isoforms together with YAP<sup>S127A</sup> phenocopies the two distinct subtypes of liver cancer. Moreover, we identify similar <i>CTNNB1</i> exon-skipping events in patients with hepatocellular carcinoma. Collectively, our findings advance our understanding of β-catenin-related tumorigenesis and reveal that CRISPR/Cas9 can be repurposed, <i>in vivo</i>, to study gain-of-function mutations of oncogenes in cancer. © 2023 The Authors. <i>The Journal of Pathology</i> published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.</p>\",\"PeriodicalId\":232,\"journal\":{\"name\":\"The Journal of Pathology\",\"volume\":\"259 4\",\"pages\":\"415-427\"},\"PeriodicalIF\":5.6000,\"publicationDate\":\"2023-01-15\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://pathsocjournals.onlinelibrary.wiley.com/doi/epdf/10.1002/path.6054\",\"citationCount\":\"1\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"The Journal of Pathology\",\"FirstCategoryId\":\"3\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/path.6054\",\"RegionNum\":2,\"RegionCategory\":\"医学\",\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"Q1\",\"JCRName\":\"ONCOLOGY\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"The Journal of Pathology","FirstCategoryId":"3","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/path.6054","RegionNum":2,"RegionCategory":"医学","ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"ONCOLOGY","Score":null,"Total":0}
引用次数: 1
引用
批量引用