In order to provide a more comprehensive, community-level understanding of marine gene flow and connectivity, we wished to first identify geographic regions of common spatial genetic divergence across multiple species along a southern temperate coastline, and then to determine which biological and ecological factors best explain the diversity in genetic patterns among species.
New Zealand (NZ) marine coastline.
Twenty-one species of benthic invertebrate.
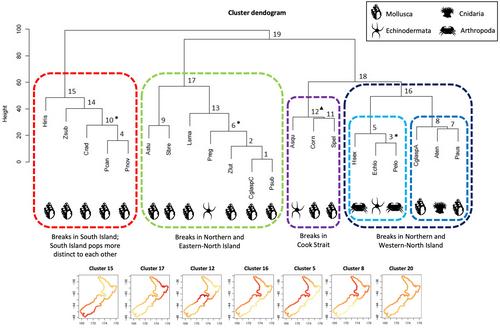
A novel approach was used that involved: (1) use of genetic divergences (F ST) from previously published studies to quantitatively describe patterns of population structure within each species as a fitted spline curve, (2) quantitatively clustering species by their similarity in geographic pattern using a dendrogram of curve similarities, and (3) then testing whether nine known life-history and ecological traits are associated with the species sharing similar genetic patterns, using distance-based regression.
Comparisons among species revealed not one, but four major common geographic patterns, within unexpected groups of species. The common locations of genetic divergence are similar to those previously identified, but differ substantially in their relative importance, compared to prior expectations. Two variables, Spawning Time and Taxon, explained significant proportions (26% and 16%) of the variation in the multivariate data.
The genogeographic clustering of population genetic divergences provided considerable insight into the concordance of marine spatial structure across species, and some potential biological drivers of those patterns. The four common patterns of population structure identified revealed that different species responded to the same environmental drivers in very different and unexpected ways. Although larval dispersal is an important factor uniting groups of species, the timing of dispersal may be more important than pelagic larval duration in NZ. These results should contribute greatly to the integration of population genetics into both community ecology and conservation management.