Hepatocellular carcinoma (HCC) represents a primary liver tumor characterized by a bleak prognosis and elevated mortality rates, yet its precise molecular mechanisms have not been fully elucidated. This study uses advanced bioinformatics techniques to discern differentially expressed genes (DEGs) implicated in the pathogenesis of HCC. The primary objective is to discover novel biomarkers and potential therapeutic targets that can contribute to the advancement of HCC research.
The bioinformatics analysis in this study primarily utilized the Gene Expression Omnibus (GEO) database as data source. Initially, the Transcriptome analysis console (TAC) screened for DEGs. Subsequently, we constructed a protein–protein interaction (PPI) network of the proteins associated to the identified DEGs with the STRING database. We obtained our hub genes using Cytoscape and confirmed the results through the GEPIA database. Furthermore, we assessed the prognostic significance of the identified hub genes using the GEPIA database. To explore the regulatory interactions, a miRNA-gene interaction network was also constructed, incorporating information from the miRDB database. For predicting the impact of gene overexpression on drug effects, we utilized CANCER DP.
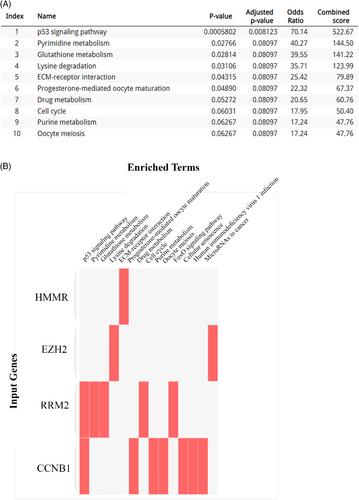
A comprehensive analysis of HCC gene expression profiles revealed a total of 4716 DEGs, consisting of 2430 upregulated genes and 2313 downregulated genes in HCC sample compared to healthy control group. These DEGs exhibited significant enrichment in key pathways such as the PI3K-Akt signaling pathway, nuclear receptors meta-pathway, and various metabolism-related pathways. Further exploration of the PPI network unveiled the P53 signaling pathway and pyrimidine metabolism as the most prominent pathways. We identified 10 hub genes (ASPM, RRM2, CCNB1, KIF14, MKI67, SHCBP1, CENPF, ANLN, HMMR, and EZH2) that exhibited significant upregulation in HCC samples compared to healthy control group. Survival analysis indicated that elevated expression levels of these genes were strongly associated with changes in overall survival in HCC patients. Lastly, we identified specific miRNAs that were found to influence the expression of these genes, providing valuable insights into potential regulatory mechanisms underlying HCC progression.
The findings of this study have successfully identified pivotal genes and pathways implicated in the pathogenesis of HCC. These novel discoveries have the potential to significantly enhance our understanding of HCC at the molecular level, opening new ways for the development of targeted therapies and improved prognosis evaluation.