Intervertebral disc degeneration (IVDD) is an age-related orthopedic degenerative disease characterized by recurrent episodes of lower back pain, the pathogenesis of which is not fully understood. This study aimed to identify key biomarkers of IVDD and its causes.
We acquired three gene expression profiles from the Gene Expression Omnibus (GEO) database, GSE56081, GSE124272, and GSE153761, and used limma fast differential analysis to identify differentially expressed genes (DEGs) between normal and IVDD samples after removing batch effects. We applied weighted gene co-expression network (WGCNA) to identify the key modular genes in GSE124272 and intersected these with DEGs. Next, A protein–protein interaction network (PPI) was constructed, and Cytoscape was used to identify the Top 10 hub genes. Functional enrichment analyses were performed using gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Three key genes were validated using Western Blot (WB) and qRT-PCR. Additionally, we predicted miRNAs involved in hub gene co-regulation and analyzed miRNA microarray data from GSE116726 to identify four differentially expressed miRNAs.
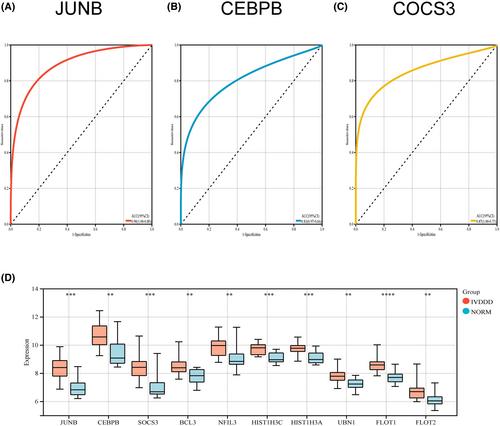
We identified 10 hub genes using bioinformatics analysis, gene function enrichment analysis revealed that they were primarily enriched in pathways, such as the TNF signaling pathway. We chose JUNB, SOCS3, and CEBPB as hub genes and used WB and qRT-PCR to confirm their expression. All three genes were overexpressed in the IVDD model group compared to the control group. Furthermore, we identified four miRNAs involved in the co-regulation of the hub genes using miRNet prediction: mir-191-5p, mir-20a-5p, mir-155-5p, and mir-124-3p. Using limma difference analysis, we discovered that mir-191-5p, mir-20a-5p, and mir-155-5p were all down-regulated and expressed in IVDD samples, but mir-124-3p showed no significant change.
JUNB, SOCS3, and CEBPB were identified as key genes in IVDD, regulated by specific miRNAs, providing potential biomarkers for early diagnosis and therapeutic targets.