Obesity-associated insulin resistance (IR) is responsible for considerable morbidity and mortality globally. Despite vast genomic data, many areas, from pathogenesis to management, still have significant knowledge gaps. We aimed to characterize visceral adipose tissue (VAT) in obesity and IR through a multi-omics approach.
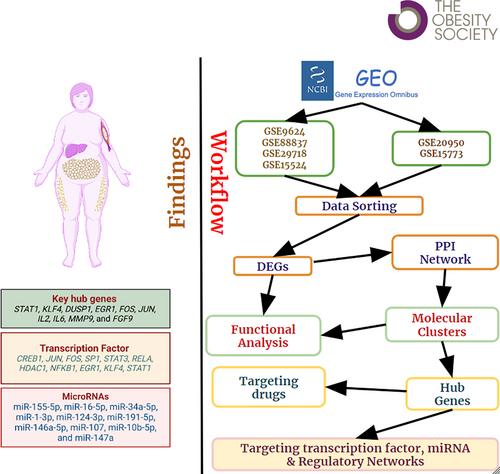
We procured data on VAT samples from the Gene Expression Omnibus (GEO) for the following two groups: 1) populations with obesity (n = 34) versus those without (n = 26); and 2) populations with obesity and IR (n = 15) versus those with obesity but without IR (n = 15). Gene set enrichment, protein-protein interaction network construction, hub gene identification, and drug-gene interactions were performed, followed by regulatory network prediction involving transcription factors (TFs) and microRNAs (miRNAs).
Interleukin signaling pathways, cellular differentiation, and regulation of immune response revealed a significant cross talk between VAT and the immune system. Other findings include cancer pathways, neurotrophin signaling, and aging. A total of 10 hub genes, i.e., STAT1, KLF4, DUSP1, EGR1, FOS, JUN, IL2, IL6, MMP9, and FGF9, 24 TFs, and approved hub gene-targeting drugs were obtained. A total of 10 targeting miRNAs (e.g., hsa-miR-155-5p, hsa-miR-34a-5p) were associated with obesity and IR-related pathways.
Our multi-omics integration method revealed hub genes, TFs, and miRNAs that can be potential targets for investigation in VAT-related inflammatory processes and IR, therapeutic management, and risk stratifications.