The composition and function of gut microbiota, lipids, and metabolites in patients with type 1 diabetes (T1D) or its association with glycemic control remains unknown. We aimed to use multi-omics sequencing technology and machine learning (ML) approaches to investigate potential function and relationships among the gut microbiota, lipids, and metabolites in T1D patients at varied glycemic levels.
We conducted a multi-omics analysis of the gut microbiome from fecal samples, metabolites, and lipids obtained from serum samples, collected from a cohort of 72 T1D patients. The patients were divided into two groups based on their hemoglobin A1c (HbA1c) levels. 16S rRNA sequencing, and metabolomics methods were applied to analyze changes in composition and function of gut microbiota, metabolites, and lipids.
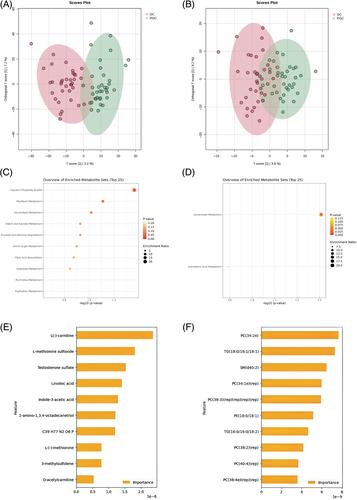
The linear discriminant analysis, Shapley additive explanations (SHAP) algorithm, and ML algorithms revealed the enrichment of Bacteroides_nordii, Bacteroides_cellulosilyticus in the glycemic control (GC) group, while Bacteroides_coprocola and Sutterella_wadsworthensis were enriched in the poor glycemic control (PGC) group. Several metabolic enrichment sets like fatty acid biosynthesis and glycerol phosphate shuttle metabolism were different between two groups. Bacteroides_nordii exhibited a negative association with D-fructose, a component involved in the starch and sucrose metabolism pathway, as well as with monoglycerides (16:0) involved in the glycerolipid metabolism pathway.
We identified distinct characteristics of gut microbiota, metabolites, and lipids in T1D patients exhibiting different levels of glycemic control. Through comprehensive analysis, microbiota (Bacteroides_nordii, Bacteroides_coprocola), metabolites (D-fructose), and lipids (Monoglycerides) may serve as potential mediators that communicated the interaction between the gut, circulatory systems, and glucose fluctuations in T1D patients.