{"title":"GEDI: An R Package for Integration of Transcriptomic Data from Multiple Platforms for Bioinformatics Applications","authors":"Mathias N. Stokholm, Maria B. Rabaglino, Haja N. Kadarmideen","doi":"10.1002/cpz1.70046","DOIUrl":null,"url":null,"abstract":"<p>Transcriptomic data is often expensive and difficult to generate in large cohorts relative to genomic data; therefore, it is often important to integrate multiple transcriptomic datasets from both microarray- and next generation sequencing (NGS)-based transcriptomic data across similar experiments or clinical trials to improve analytical power and discovery of novel transcripts and genes. However, transcriptomic data integration presents a few challenges including reannotation and batch effect removal. We developed the Gene Expression Data Integration (GEDI) R package to enable transcriptomic data integration by combining existing R packages. With just four functions, the GEDI R package makes constructing a transcriptomic data integration pipeline straightforward. Together, the functions overcome the complications in transcriptomic data integration by automatically reannotating the data and removing the batch effect. The removal of the batch effect is verified with principal component analysis and the data integration is verified using a logistic regression model with forward stepwise feature selection. To demonstrate the functionalities of the GEDI package, we integrated five bovine endometrial transcriptomic datasets from the NCBI Gene Expression Omnibus. These transcriptomic datasets were from multiple high-throughput platforms, namely, array-based Affymetrix and Agilent platforms, and NGS-based Illumina paired-end RNA-seq platform. Furthermore, we compared the GEDI package to existing tools and found that GEDI is the only tool that provides a full transcriptomic data integration pipeline including verification of both batch effect removal and data integration for downstream genomic and bioinformatics applications. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: ReadGE, a function to import gene expression datasets</p><p><b>Basic Protocol 2</b>: GEDI, a function to reannotate and merge gene expression datasets</p><p><b>Basic Protocol 3</b>: BatchCorrection, a function to remove batch effects from gene expression data</p><p><b>Basic Protocol 4</b>: VerifyGEDI, a function to confirm successful integration of gene expression data</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 10","pages":""},"PeriodicalIF":2.2000,"publicationDate":"2024-10-25","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70046","citationCount":"0","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/cpz1.70046","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
Abstract
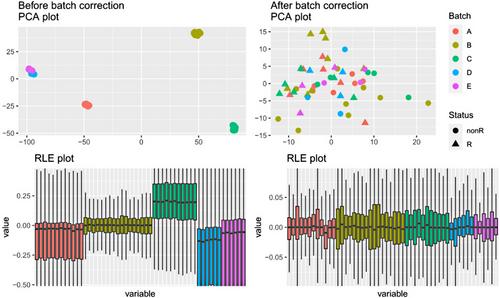
Transcriptomic data is often expensive and difficult to generate in large cohorts relative to genomic data; therefore, it is often important to integrate multiple transcriptomic datasets from both microarray- and next generation sequencing (NGS)-based transcriptomic data across similar experiments or clinical trials to improve analytical power and discovery of novel transcripts and genes. However, transcriptomic data integration presents a few challenges including reannotation and batch effect removal. We developed the Gene Expression Data Integration (GEDI) R package to enable transcriptomic data integration by combining existing R packages. With just four functions, the GEDI R package makes constructing a transcriptomic data integration pipeline straightforward. Together, the functions overcome the complications in transcriptomic data integration by automatically reannotating the data and removing the batch effect. The removal of the batch effect is verified with principal component analysis and the data integration is verified using a logistic regression model with forward stepwise feature selection. To demonstrate the functionalities of the GEDI package, we integrated five bovine endometrial transcriptomic datasets from the NCBI Gene Expression Omnibus. These transcriptomic datasets were from multiple high-throughput platforms, namely, array-based Affymetrix and Agilent platforms, and NGS-based Illumina paired-end RNA-seq platform. Furthermore, we compared the GEDI package to existing tools and found that GEDI is the only tool that provides a full transcriptomic data integration pipeline including verification of both batch effect removal and data integration for downstream genomic and bioinformatics applications. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1: ReadGE, a function to import gene expression datasets
Basic Protocol 2: GEDI, a function to reannotate and merge gene expression datasets
Basic Protocol 3: BatchCorrection, a function to remove batch effects from gene expression data
Basic Protocol 4: VerifyGEDI, a function to confirm successful integration of gene expression data

GEDI:为生物信息学应用整合来自多个平台的转录组数据的 R 软件包。
与基因组数据相比,转录组数据通常成本高昂,而且难以在大型队列中生成;因此,通常需要整合类似实验或临床试验中基于微阵列和新一代测序(NGS)的多个转录组数据集,以提高分析能力并发现新的转录本和基因。然而,转录组数据整合面临着一些挑战,包括重新注释和批次效应去除。我们开发了基因表达数据整合(GEDI)R软件包,通过结合现有的R软件包实现转录组数据整合。GEDI R软件包仅有四个函数,使构建转录组数据整合管道变得简单易行。这些函数通过自动重新标注数据和消除批次效应,克服了转录组数据整合的复杂性。批次效应的消除通过主成分分析进行了验证,数据整合则通过带有前向逐步特征选择的逻辑回归模型进行了验证。为了展示 GEDI 软件包的功能,我们整合了 NCBI 基因表达总库中的五个牛子宫内膜转录组数据集。这些转录组数据集来自多个高通量平台,即基于阵列的 Affymetrix 和 Agilent 平台,以及基于 NGS 的 Illumina 成对端 RNA-seq 平台。此外,我们还将 GEDI 软件包与现有工具进行了比较,发现 GEDI 是唯一能提供完整转录组数据整合流水线的工具,包括批次效应去除验证和下游基因组学与生物信息学应用的数据整合。© 2024 作者。当前协议》由 Wiley Periodicals LLC 出版。基本协议 1:ReadGE,一个导入基因表达数据集的函数 基本协议 2:GEDI,一个重新标注和合并基因表达数据集的函数 基本协议 3:BatchCorrection,一个从基因表达数据中去除批次效应的函数 基本协议 4:VerifyGEDI,一个确认基因表达数据成功整合的函数。
本文章由计算机程序翻译,如有差异,请以英文原文为准。



