{"title":"The TELCoMB Protocol for High-Sensitivity Detection of ARG-MGE Colocalizations in Complex Microbial Communities","authors":"Jonathan E. Bravo, Ilya Slizovskiy, Nathalie Bonin, Marco Oliva, Noelle Noyes, Christina Boucher","doi":"10.1002/cpz1.70031","DOIUrl":null,"url":null,"abstract":"<p>Understanding the genetic basis of antimicrobial resistance is crucial for developing effective mitigation strategies. One necessary step is to identify the antimicrobial resistance genes (ARGs) within a microbial population, referred to as the resistome, as well as the mobile genetic elements (MGEs) harboring ARGs. Although shotgun metagenomics has been successful in detecting ARGs and MGEs within a microbiome, it is limited by low sensitivity. Enrichment using cRNA biotinylated probes has been applied to address this limitation, enhancing the detection of rare ARGs and MGEs, especially when combined with long-read sequencing. Here, we present the TELCoMB protocol, a Snakemake workflow that elucidates resistome and mobilome composition and diversity and uncovers ARG-MGE colocalizations. The protocol supports both short- and long-read sequencing and does not require enrichment, making it versatile for various genomic data types. TELCoMB generates publication-ready figures and CSV files for comprehensive analysis, improving our understanding of antimicrobial resistance mechanisms and spread. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Installing TELCOMB Locally</p><p><b>Alternate Protocol</b>: Installing TELCOMB on a SLURM Cluster</p><p><b>Basic Protocol 2</b>: Data Preprocessing</p><p><b>Basic Protocol 3</b>: Calculation of Resistome Distribution and Composition</p><p><b>Basic Protocol 4</b>: Identification of ARG-MGE Colocalizations</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 10","pages":""},"PeriodicalIF":2.2000,"publicationDate":"2024-10-24","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70031","citationCount":"0","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/cpz1.70031","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
Abstract
Understanding the genetic basis of antimicrobial resistance is crucial for developing effective mitigation strategies. One necessary step is to identify the antimicrobial resistance genes (ARGs) within a microbial population, referred to as the resistome, as well as the mobile genetic elements (MGEs) harboring ARGs. Although shotgun metagenomics has been successful in detecting ARGs and MGEs within a microbiome, it is limited by low sensitivity. Enrichment using cRNA biotinylated probes has been applied to address this limitation, enhancing the detection of rare ARGs and MGEs, especially when combined with long-read sequencing. Here, we present the TELCoMB protocol, a Snakemake workflow that elucidates resistome and mobilome composition and diversity and uncovers ARG-MGE colocalizations. The protocol supports both short- and long-read sequencing and does not require enrichment, making it versatile for various genomic data types. TELCoMB generates publication-ready figures and CSV files for comprehensive analysis, improving our understanding of antimicrobial resistance mechanisms and spread. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1: Installing TELCOMB Locally
Alternate Protocol: Installing TELCOMB on a SLURM Cluster
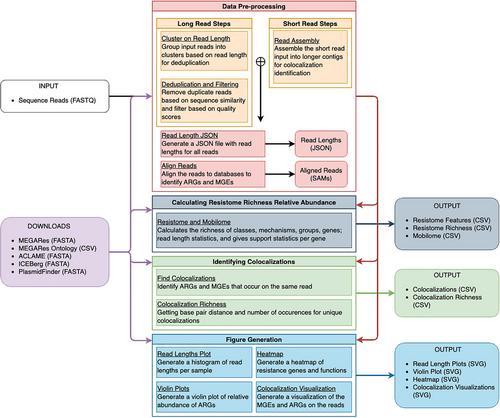
Basic Protocol 2: Data Preprocessing
Basic Protocol 3: Calculation of Resistome Distribution and Composition
Basic Protocol 4: Identification of ARG-MGE Colocalizations

高灵敏度检测复杂微生物群落中 ARG-MGE 共定位的 TELCoMB 方案。
了解抗菌素耐药性的遗传基础对于制定有效的缓解策略至关重要。其中一个必要步骤是确定微生物种群中的抗菌药耐药性基因(ARGs)(称为耐药性组)以及携带 ARGs 的移动遗传因子(MGEs)。尽管霰弹枪元基因组学已成功检测出微生物群中的 ARGs 和 MGEs,但其灵敏度较低。使用 cRNA 生物素化探针进行富集可以解决这一局限性,从而提高对罕见 ARGs 和 MGEs 的检测能力,尤其是在与长线程测序相结合时。在这里,我们介绍一种Snakemake工作流程--TELCoMB协议,它能阐明抗性组和动员组的组成和多样性,并发现ARG-MGE共定位。该方案支持短线程和长线程测序,无需富集,因此适用于各种基因组数据类型。TELCoMB 可生成可供发表的图表和 CSV 文件,用于综合分析,从而提高我们对抗菌药耐药性机制和传播的认识。© 2024 作者。当前协议由 Wiley Periodicals LLC 出版。基本协议 1:在本地安装 TELCOMB 备选协议:基本协议 2:数据预处理 基本协议 3:抗性组分布和组成的计算 基本协议 4:ARG-MGE 共定位的识别。
本文章由计算机程序翻译,如有差异,请以英文原文为准。



