Species presence–absence data can be time-consuming and logistically difficult to obtain across large spatial extents. Yet these data are important for ensuring changes in species distributions are accurately monitored and are vital for ensuring appropriate conservation actions are undertaken. Here, we demonstrate how environmental DNA (eDNA) sampling can be used to systematically collect species occupancy data rapidly and efficiently across vast spatial domains to improve understanding of factors influencing species distributions.
South-eastern Australia.
We use a widely distributed, but near-threatened species, the platypus (Ornithorhynchus anatinus), as a test case and undertake an environmentally stratified systematic survey to assess the presence–absence of platypus eDNA at 504 sites across 584,292 km2 of south-eastern Australia, representing ~37% of the species' extensive distribution. Site occupancy-detection models were used to analyse how landscape- and site-level factors affect platypus occupancy, enabling us to incorporate uncertainty at the different levels inherent in eDNA sampling (site, water sample replicate and qPCR replicate).
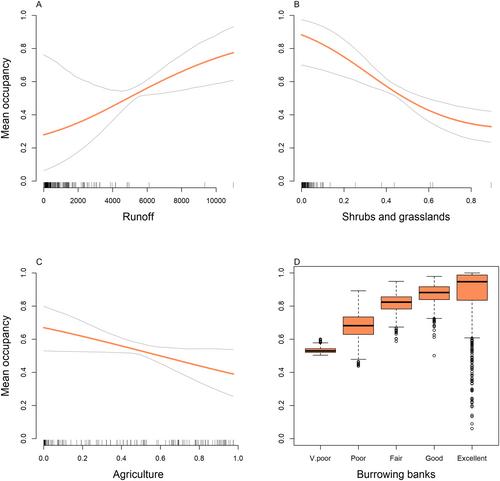
Platypus eDNA was detected at 272 sites (~54%) with platypuses more likely to occupy sites in catchments with increased runoff and less zero-flow days, and sites with access to banks suitable for burrowing. Platypuses were less likely to occupy sites in catchments with a high proportion of shrubs and grasslands, or agricultural land use.
These data provide an important large-scale validation of the landscape- and site-level factors influencing platypus occupancy that can be used to inform future conservation efforts. Our case study shows that systematically designed, stratified eDNA surveys provide an efficient means to understand how environmental characteristics affect species occupancy across broad environmental gradients. The methods employed here can be applied to aquatic and semi-aquatic species globally, providing unprecedented opportunities to understand biodiversity status and change and provide insights for current and future conservation actions.