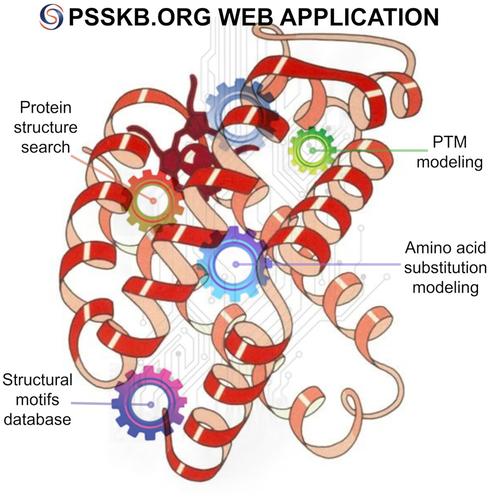
The proteins expressed during the cell cycle determine cell function and ensure signaling pathway activation in response to environmental influences. Developments in structural biology, biophysics, and bioinformatics provide information on the structure and function of particular proteins including that on the structural changes in proteins due to post-translational modification (PTM) and amino acid substitutions (AAS), which is essential for understanding protein function and life cycle. These are PTMs and AASs that often modulate the function and alter the stability and localization of a protein in a cell. PSSKB is a platform that integrates all necessary tools for modeling the five common natural modifications and all canonical AASs in proteins. The available tools are not limited to the local database, so the user can select a protein from Uniprot ID or PDB ID. The result will be a three-dimensional (3D) representation of the modified structure, as well as an analysis of the changes in the performance of the intact and modified structures after energy minimization compared with the original structure, which not only makes it possible to evaluate AAS/PTM influence of on a protein's characteristics but also to use the 3D model for further studies. Additionally, PSSKB enables the user to search, align, overlay, and determine the exact coordinates of protein structure fragments. The search results are a set of structural motifs similar to the query and ranked by statistical significance. The platform is fully functional and publicly available at https://psskb.org/. No registration is required to access the platform. A tutorial video can be found at https://psskb.org/page/about. Services provided on the platform are based on previously developed and published software. SCPacker applied for PTM Modeling and AAS services available at GitHub (https://github.com/protdb/SCPacker). SaFoldNet applied for a Similar Search service is also available at GitHub (https://github.com/protdb/ABBNet).