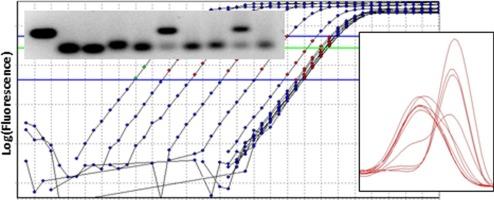
Quantitative PCR allows the precise measurement of DNA concentrations and is generally considered to be straightforward and trouble free. However, a survey with 93 validated assays for genes in the Wnt-pathway showed that the amplification of nonspecific products occurs frequently and is unrelated to Cq or PCR efficiency values. Titration experiments showed that the occurrence of low and high melting temperature artifacts was shown to be determined by annealing temperature, primer concentration and cDNA input. To explore the range of input variations that occur in the normal use of the Cre assay these conditions were mimicked in a complete two-way design of template −plasmid DNA- and non-template −mouse cDNA- concentrations. These experiments showed that the frequency of the amplification of the correct product and the artifact, as well as the valid quantification of the correct product, depended on the concentration of the non-template cDNA. This finding questions the interpretation of dilution series in which template as well as non-template concentrations are simultaneously decreasing. Repetition of this cDNA concentration experiment with other templates revealed that exact reproduction qPCR experiments was affected by the time it takes to complete the pipetting of a qPCR plate. Long bench times were observed to lead to significantly more artifacts. However, the measurement of artifact-associated fluorescence can be avoided by inclusion of a small heating step after the elongation phase in the amplification protocol. Taken together, this trouble-shooting journey showed that reliability and reproducibility of qPCR experiments not only depends on standardization and reporting of the biochemistry and technical aspects but also on hitherto neglected factors as sample dilution and waiting times in the laboratory work flow.