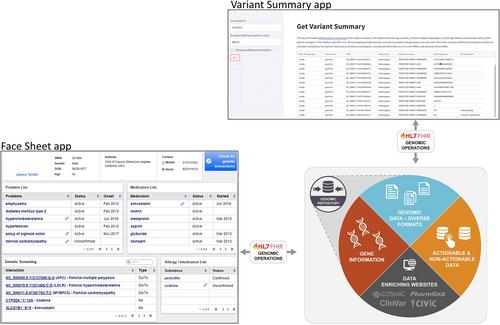
Variant annotation is a critical component in next-generation sequencing, enabling a sequencing lab to comb through a sea of variants in order to hone in on those likely to be most significant, and providing clinicians with necessary context for decision-making. But with the rapid evolution of genomics knowledge, reported annotations can quickly become out-of-date. Under the ONC Sync for Genes program, our team sought to standardize the sharing of dynamically annotated variants (e.g., variants annotated on demand, based on current knowledge). The computable biomedical knowledge artifacts that were developed enable a clinical decision support (CDS) application to surface up-to-date annotations to clinicians.
The work reported in this article relies on the Health Level 7 Fast Healthcare Interoperability Resources (FHIR) Genomics and Global Alliance for Genomics and Health (GA4GH) Variant Annotation (VA) standards. We developed a CDS pipeline that dynamically annotates patient's variants through an intersection with current knowledge and serves up the FHIR-encoded variants and annotations (diagnostic and therapeutic implications, molecular consequences, population allele frequencies) via FHIR Genomics Operations. ClinVar, CIViC, and PharmGKB were used as knowledge sources, encoded as per the GA4GH VA specification.
Primary public artifacts from this project include a GitHub repository with all source code, a Swagger interface that allows anyone to visualize and interact with the code using only a web browser, and a backend database where all (synthetic and anonymized) patient data and knowledge are housed.
We found that variant annotation varies in complexity based on the variant type, and that various bioinformatics strategies can greatly improve automated annotation fidelity. More importantly, we demonstrated the feasibility of an ecosystem where genomic knowledge bases have standardized knowledge (e.g., based on the GA4GH VA spec), and CDS applications can dynamically leverage that knowledge to provide real-time decision support, based on current knowledge, to clinicians at the point of care.