Like most other cell surface proteins, α5β1 integrin is glycosylated, which is required for its various activities in ways that mostly remain to be determined.
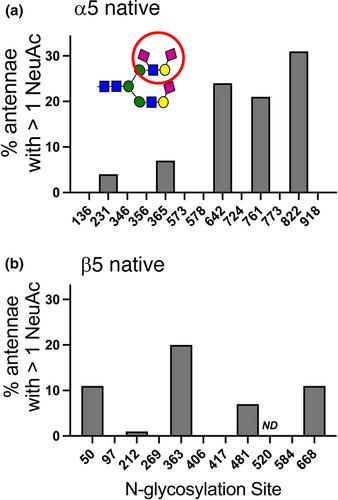
Here, we have established the first comprehensive site-specific glycan map of α5β1 integrin that was purified from a natural source, that is, rat liver. This analysis revealed striking site selective variations in glycan composition. Complex bi, tri, or tetraantennary N-glycans were predominant at various proportions at most potential N-glycosylation sites. A few of these sites were nonglycosylated or contained high mannose or hybrid glycans, indicating that early N-glycan processing was hindered. Almost all complex N-glycans had fully galactosylated and sialylated antennae. Moderate levels of core fucosylation and high levels of O-acetylation of NeuAc residues were observed at certain sites. An O-linked HexNAc was found in an EGF-like domain of β1 integrin. The extensive glycan information that results from our study was projected onto a map of α5β1 integrin that was obtained by homology modeling. We have used this model for the discussion of how glycosylation might be used in the functional cycle of α5β1 integrin. A striking example concerns the involvement of glycan-binding galectins in the regulation of the molecular homeostasis of glycoproteins at the cell surface through the formation of lattices or endocytic pits according to the glycolipid-lectin (GL-Lect) hypothesis.
We expect that the glycoproteomics data of the current study will serve as a resource for the exploration of structural mechanisms by which glycans control α5β1 integrin activity and endocytic trafficking.
Glycosylation of α5β1 integrin has been implicated in multiple aspects of integrin function and structure. Yet, detailed knowledge of its glycosylation, notably the specific sites of glycosylation, is lacking. Furthermore, the α5β1 integrin preparation that was analyzed here is from a natural source, which is of importance as there is not a lot of literature in the field about the glycosylation of “native” glycoproteins.