Anadromous Striped Bass Morone saxatilis are dominant predators in estuaries and coastal areas along the U.S. Atlantic coast, with the potential to exert top-down control on prey populations. Although Striped Bass diets have been studied previously, spatiotemporal patterns of diet across ontogeny remain poorly understood, especially for young fish in shallow nursery habitats.
We collected and examined gut contents from adult, juvenile, and young-of-year (age-0) Striped Bass from nine rivers across the Maryland and Virginia portions of Chesapeake Bay during summer and fall 2018. We compared the use of traditional morphological inspection and new amplicon-based next-generation sequencing methods for identifying gut contents.
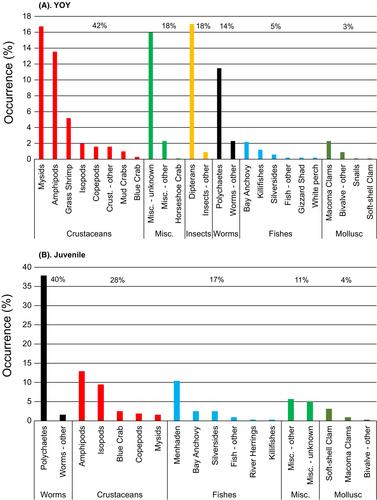
Striped Bass in shallow tributary habitats of Chesapeake Bay had diverse diets that varied strongly with ontogeny and salinity zone. In particular, the diet of age-0 Striped Bass varied greatly from those of juveniles and adults when age-0 fish foraged in freshwater habitats. Although our results on prey consumed aligned with previous surveys, we identified additional taxa as important prey for these young fish, including dipteran insects, Banded Killifish Fundulus diaphanus, Inland Silverside Menidia beryllina, bay barnacle Amphibalanus improvisus, and grass shrimp Palaemon spp. Comparison of methodologies indicated that 40% of prey by weight could not be identified with morphological analysis, while 76% of mitochondrial cytochrome oxidase I sequences could be assigned binomial names, allowing for high-resolution taxonomic comparisons.
This study adds to the growing body of evidence that amplicon-based next-generation sequencing methods are far superior to traditional morphological analyses of gut contents for fine-scale taxonomic resolution of prey.