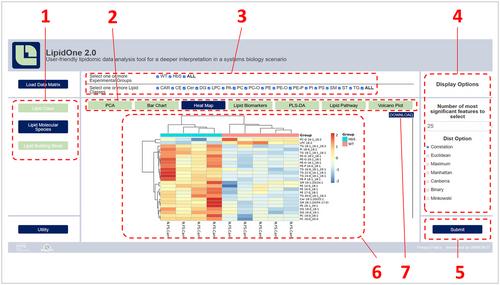
{"title":"LipidOne 2.0:发现脂质体数据中隐藏的生物学意义的网络工具","authors":"Husam B. R. Alabed, Dorotea Frongia Mancini, Sandra Buratta, Eleonora Calzoni, Danika Di Giacomo, Carla Emiliani, Sabata Martino, Lorena Urbanelli, Roberto Maria Pellegrino","doi":"10.1002/cpz1.70009","DOIUrl":null,"url":null,"abstract":"<p>LipidOne 2.0 (https://lipidone.eu) is a new web bioinformatic tool for the analysis of lipidomic data. It facilitates the exploration of the three structural levels of lipids: classes, molecular species, and lipid building blocks (acyl, alkyl, or alkenes chains). The tool's flexibility empowers users to seamlessly include or exclude experimental groups and lipid classes at any stage of the analysis. LipidOne 2.0 offers a range of mono- and multivariate statistical analyses, specifically tailored to each structural level. This includes a novel lipid biomarker identification function, integrating four diverse statistical parameters. LipidOne 2.0 incorporates Lipid Pathway analysis across all three structural levels of lipids. Users can identify lipid-involved reactions through case-control comparisons, generating lists of genes/enzymes and their activation states based on <i>Z</i> scores. Accessible without the need for registration, LipidOne 2.0 provides a user-friendly and efficient platform for exploring and analyzing lipidomic data. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Dataset preparation for LipidOne 2.0</p><p><b>Support Protocol</b>: Lipid nomenclature from spectrometric experiments</p><p><b>Basic Protocol 2</b>: Uploading a dataset into LipidOne 2.0</p><p><b>Basic Protocol 3</b>: Data mining of lipidomic dataset by LipidOne 2.0</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 9","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2024-09-20","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70009","citationCount":"0","resultStr":"{\"title\":\"LipidOne 2.0: A Web Tool for Discovering Biological Meanings Hidden in Lipidomic Data\",\"authors\":\"Husam B. R. Alabed, Dorotea Frongia Mancini, Sandra Buratta, Eleonora Calzoni, Danika Di Giacomo, Carla Emiliani, Sabata Martino, Lorena Urbanelli, Roberto Maria Pellegrino\",\"doi\":\"10.1002/cpz1.70009\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>LipidOne 2.0 (https://lipidone.eu) is a new web bioinformatic tool for the analysis of lipidomic data. It facilitates the exploration of the three structural levels of lipids: classes, molecular species, and lipid building blocks (acyl, alkyl, or alkenes chains). The tool's flexibility empowers users to seamlessly include or exclude experimental groups and lipid classes at any stage of the analysis. LipidOne 2.0 offers a range of mono- and multivariate statistical analyses, specifically tailored to each structural level. This includes a novel lipid biomarker identification function, integrating four diverse statistical parameters. LipidOne 2.0 incorporates Lipid Pathway analysis across all three structural levels of lipids. Users can identify lipid-involved reactions through case-control comparisons, generating lists of genes/enzymes and their activation states based on <i>Z</i> scores. Accessible without the need for registration, LipidOne 2.0 provides a user-friendly and efficient platform for exploring and analyzing lipidomic data. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Dataset preparation for LipidOne 2.0</p><p><b>Support Protocol</b>: Lipid nomenclature from spectrometric experiments</p><p><b>Basic Protocol 2</b>: Uploading a dataset into LipidOne 2.0</p><p><b>Basic Protocol 3</b>: Data mining of lipidomic dataset by LipidOne 2.0</p>\",\"PeriodicalId\":93970,\"journal\":{\"name\":\"Current protocols\",\"volume\":\"4 9\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2024-09-20\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70009\",\"citationCount\":\"0\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current protocols\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.70009\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"\",\"JCRName\":\"\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.70009","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0