{"title":"记录和解读诱导多能干细胞衍生心肌细胞的活性钙瞬态","authors":"Maedeh Mozneb, Jemima Moses, Madelyn Arzt, Sean Escopete, Arun Sharma","doi":"10.1002/cpz1.70030","DOIUrl":null,"url":null,"abstract":"<p>Calcium plays a pivotal role in the excitation-contraction coupling process in cardiomyocytes, a critical multi-parametric event leading to rhythmic contraction. Over the past few decades, calcium signaling in cardiomyocytes has been extensively studied in cardiovascular sciences. However, a standard methodology is needed not only to trace the calcium within cells but also to remove signal processing biases and to accurately interpret the features of calcium transient signals in relation to cardiomyocyte electrophysiology. This article outlines the use of genetically encoded calcium indicator (GCaMP) human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) to record calcium transients. These cells express a green fluorescent signal when calcium binds to intracellular calmodulin, a key regulator of calcium signaling. The extraction and processing of calcium transient waveforms are performed using ImageJ and MATLAB software. Key features of these waveforms are then identified and categorized based on their physiological relevance to cardiomyocyte function. Additionally, this work includes a Support Protocol for the successful replating of cardiomyocytes onto non-traditional culture platforms, such as metallic sensors and polymer-based substrates, to facilitate data multiplexing. The three Basic Protocols outlined here provide a comprehensive approach for maintaining, expanding, and differentiating the GCaMP hiPSCs, video recording of calcium transients, and the subsequent signal extraction, preprocessing, analysis, and data visualization. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Maintenance, expansion, and differentiation of genetically encoded calcium indicator hiPSCs</p><p><b>Support Protocol</b>: Replating GCaMP hiPSC-CMs for stimulation and multielectrode array studies</p><p><b>Basic Protocol 2</b>: Video recording from calcium transients of GCaMP hiPSC-CMs</p><p><b>Basic Protocol 3</b>: Signal extraction, preprocessing, analysis, and data visualization</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 10","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2024-10-24","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70030","citationCount":"0","resultStr":"{\"title\":\"Recording and Interpretation of Active Calcium Transients in Induced Pluripotent Stem Cell-Derived Cardiomyocytes\",\"authors\":\"Maedeh Mozneb, Jemima Moses, Madelyn Arzt, Sean Escopete, Arun Sharma\",\"doi\":\"10.1002/cpz1.70030\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>Calcium plays a pivotal role in the excitation-contraction coupling process in cardiomyocytes, a critical multi-parametric event leading to rhythmic contraction. Over the past few decades, calcium signaling in cardiomyocytes has been extensively studied in cardiovascular sciences. However, a standard methodology is needed not only to trace the calcium within cells but also to remove signal processing biases and to accurately interpret the features of calcium transient signals in relation to cardiomyocyte electrophysiology. This article outlines the use of genetically encoded calcium indicator (GCaMP) human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) to record calcium transients. These cells express a green fluorescent signal when calcium binds to intracellular calmodulin, a key regulator of calcium signaling. The extraction and processing of calcium transient waveforms are performed using ImageJ and MATLAB software. Key features of these waveforms are then identified and categorized based on their physiological relevance to cardiomyocyte function. Additionally, this work includes a Support Protocol for the successful replating of cardiomyocytes onto non-traditional culture platforms, such as metallic sensors and polymer-based substrates, to facilitate data multiplexing. The three Basic Protocols outlined here provide a comprehensive approach for maintaining, expanding, and differentiating the GCaMP hiPSCs, video recording of calcium transients, and the subsequent signal extraction, preprocessing, analysis, and data visualization. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Maintenance, expansion, and differentiation of genetically encoded calcium indicator hiPSCs</p><p><b>Support Protocol</b>: Replating GCaMP hiPSC-CMs for stimulation and multielectrode array studies</p><p><b>Basic Protocol 2</b>: Video recording from calcium transients of GCaMP hiPSC-CMs</p><p><b>Basic Protocol 3</b>: Signal extraction, preprocessing, analysis, and data visualization</p>\",\"PeriodicalId\":93970,\"journal\":{\"name\":\"Current protocols\",\"volume\":\"4 10\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2024-10-24\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70030\",\"citationCount\":\"0\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current protocols\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.70030\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"\",\"JCRName\":\"\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.70030","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
Recording and Interpretation of Active Calcium Transients in Induced Pluripotent Stem Cell-Derived Cardiomyocytes
Calcium plays a pivotal role in the excitation-contraction coupling process in cardiomyocytes, a critical multi-parametric event leading to rhythmic contraction. Over the past few decades, calcium signaling in cardiomyocytes has been extensively studied in cardiovascular sciences. However, a standard methodology is needed not only to trace the calcium within cells but also to remove signal processing biases and to accurately interpret the features of calcium transient signals in relation to cardiomyocyte electrophysiology. This article outlines the use of genetically encoded calcium indicator (GCaMP) human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) to record calcium transients. These cells express a green fluorescent signal when calcium binds to intracellular calmodulin, a key regulator of calcium signaling. The extraction and processing of calcium transient waveforms are performed using ImageJ and MATLAB software. Key features of these waveforms are then identified and categorized based on their physiological relevance to cardiomyocyte function. Additionally, this work includes a Support Protocol for the successful replating of cardiomyocytes onto non-traditional culture platforms, such as metallic sensors and polymer-based substrates, to facilitate data multiplexing. The three Basic Protocols outlined here provide a comprehensive approach for maintaining, expanding, and differentiating the GCaMP hiPSCs, video recording of calcium transients, and the subsequent signal extraction, preprocessing, analysis, and data visualization. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
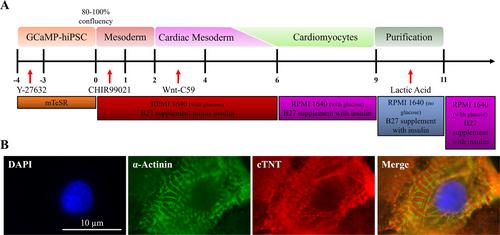
Basic Protocol 1: Maintenance, expansion, and differentiation of genetically encoded calcium indicator hiPSCs
Support Protocol: Replating GCaMP hiPSC-CMs for stimulation and multielectrode array studies
Basic Protocol 2: Video recording from calcium transients of GCaMP hiPSC-CMs
Basic Protocol 3: Signal extraction, preprocessing, analysis, and data visualization