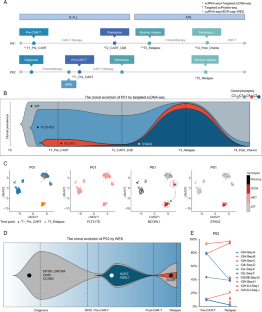
非 KMT2A 重排 B-ALL 患者在 CD19 CAR-T 治疗下的系谱转换动态演变
Shaowei Qiu, Yihan Mei, Runxia Gu, Yu Liu, Manling Chen, Haiyan Xing, Kejing Tang, Zheng Tian, Qing Rao, Donglin Yang, Aiming Pang, Shuning Wei, Yujiao Jia, Huijun Wang, Sizhou Feng, Hui Wei, Ping Zhu, Min Wang, Ying Wang, Wenbing Liu, Jianxiang Wang
求助PDF
{"title":"非 KMT2A 重排 B-ALL 患者在 CD19 CAR-T 治疗下的系谱转换动态演变","authors":"Shaowei Qiu, Yihan Mei, Runxia Gu, Yu Liu, Manling Chen, Haiyan Xing, Kejing Tang, Zheng Tian, Qing Rao, Donglin Yang, Aiming Pang, Shuning Wei, Yujiao Jia, Huijun Wang, Sizhou Feng, Hui Wei, Ping Zhu, Min Wang, Ying Wang, Wenbing Liu, Jianxiang Wang","doi":"10.1038/s41375-024-02449-7","DOIUrl":null,"url":null,"abstract":"","PeriodicalId":18109,"journal":{"name":"Leukemia","volume":"39 1","pages":"238-242"},"PeriodicalIF":12.8000,"publicationDate":"2024-10-31","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"","citationCount":"0","resultStr":"{\"title\":\"The dynamic evolution of lineage switch under CD19 CAR-T treatment in non-KMT2A rearranged B-ALL patients\",\"authors\":\"Shaowei Qiu, Yihan Mei, Runxia Gu, Yu Liu, Manling Chen, Haiyan Xing, Kejing Tang, Zheng Tian, Qing Rao, Donglin Yang, Aiming Pang, Shuning Wei, Yujiao Jia, Huijun Wang, Sizhou Feng, Hui Wei, Ping Zhu, Min Wang, Ying Wang, Wenbing Liu, Jianxiang Wang\",\"doi\":\"10.1038/s41375-024-02449-7\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"\",\"PeriodicalId\":18109,\"journal\":{\"name\":\"Leukemia\",\"volume\":\"39 1\",\"pages\":\"238-242\"},\"PeriodicalIF\":12.8000,\"publicationDate\":\"2024-10-31\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"\",\"citationCount\":\"0\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Leukemia\",\"FirstCategoryId\":\"3\",\"ListUrlMain\":\"https://www.nature.com/articles/s41375-024-02449-7\",\"RegionNum\":1,\"RegionCategory\":\"医学\",\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"Q1\",\"JCRName\":\"HEMATOLOGY\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Leukemia","FirstCategoryId":"3","ListUrlMain":"https://www.nature.com/articles/s41375-024-02449-7","RegionNum":1,"RegionCategory":"医学","ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"HEMATOLOGY","Score":null,"Total":0}
引用次数: 0
引用
批量引用