下载PDF
{"title":"使用Mobile-CRISPRi在多种细菌中可编程基因敲除","authors":"Amy B. Banta, Ryan D. Ward, Jennifer S. Tran, Emily E. Bacon, Jason M. Peters","doi":"10.1002/cpmc.130","DOIUrl":null,"url":null,"abstract":"<p>Facile bacterial genome sequencing has unlocked a veritable treasure trove of novel genes awaiting functional exploration. To make the most of this opportunity requires powerful genetic tools that can target all genes in diverse bacteria. CRISPR interference (CRISPRi) is a programmable gene-knockdown tool that uses an RNA-protein complex comprised of a single guide RNA (sgRNA) and a catalytically inactive Cas9 nuclease (dCas9) to sterically block transcription of target genes. We previously developed a suite of modular CRISPRi systems that transfer by conjugation and integrate into the genomes of diverse bacteria, which we call Mobile-CRISPRi. Here, we provide detailed protocols for the modification and transfer of Mobile-CRISPRi vectors for the purpose of knocking down target genes in bacteria of interest. We further discuss strategies for optimizing Mobile-CRISPRi knockdown, transfer, and integration. We cover the following basic protocols: sgRNA design, cloning new sgRNA spacers into Mobile-CRISPRi vectors, Tn<i>7</i> transfer of Mobile-CRISPRi to Gram-negative bacteria, and ICE<i>Bs1</i> transfer of Mobile-CRISPRi to Bacillales. © 2020 The Authors.</p><p><b>Basic Protocol 1</b>: sgRNA design</p><p><b>Basic Protocol 2</b>: Cloning of new sgRNA spacers into Mobile-CRISPRi vectors</p><p><b>Basic Protocol 3</b>: Tn<i>7</i> transfer of Mobile-CRISPRi to Gram-negative bacteria</p><p><b>Basic Protocol 4</b>: ICE<i>Bs1</i> transfer of Mobile-CRISPRi to Bacillales</p><p><b>Support Protocol 1</b>: Quantification of CRISPRi repression using fluorescent reporters</p><p><b>Support Protocol 2</b>: Testing for gene essentiality using CRISPRi spot assays on plates</p><p><b>Support Protocol 3</b>: Transformation of <i>E. coli</i> by electroporation</p><p><b>Support Protocol 4</b>: Transformation of CaCl<sub>2</sub>-competent <i>E. coli</i></p>","PeriodicalId":39967,"journal":{"name":"Current Protocols in Microbiology","volume":"59 1","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2020-12-17","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cpmc.130","citationCount":"7","resultStr":"{\"title\":\"Programmable Gene Knockdown in Diverse Bacteria Using Mobile-CRISPRi\",\"authors\":\"Amy B. Banta, Ryan D. Ward, Jennifer S. Tran, Emily E. Bacon, Jason M. Peters\",\"doi\":\"10.1002/cpmc.130\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>Facile bacterial genome sequencing has unlocked a veritable treasure trove of novel genes awaiting functional exploration. To make the most of this opportunity requires powerful genetic tools that can target all genes in diverse bacteria. CRISPR interference (CRISPRi) is a programmable gene-knockdown tool that uses an RNA-protein complex comprised of a single guide RNA (sgRNA) and a catalytically inactive Cas9 nuclease (dCas9) to sterically block transcription of target genes. We previously developed a suite of modular CRISPRi systems that transfer by conjugation and integrate into the genomes of diverse bacteria, which we call Mobile-CRISPRi. Here, we provide detailed protocols for the modification and transfer of Mobile-CRISPRi vectors for the purpose of knocking down target genes in bacteria of interest. We further discuss strategies for optimizing Mobile-CRISPRi knockdown, transfer, and integration. We cover the following basic protocols: sgRNA design, cloning new sgRNA spacers into Mobile-CRISPRi vectors, Tn<i>7</i> transfer of Mobile-CRISPRi to Gram-negative bacteria, and ICE<i>Bs1</i> transfer of Mobile-CRISPRi to Bacillales. © 2020 The Authors.</p><p><b>Basic Protocol 1</b>: sgRNA design</p><p><b>Basic Protocol 2</b>: Cloning of new sgRNA spacers into Mobile-CRISPRi vectors</p><p><b>Basic Protocol 3</b>: Tn<i>7</i> transfer of Mobile-CRISPRi to Gram-negative bacteria</p><p><b>Basic Protocol 4</b>: ICE<i>Bs1</i> transfer of Mobile-CRISPRi to Bacillales</p><p><b>Support Protocol 1</b>: Quantification of CRISPRi repression using fluorescent reporters</p><p><b>Support Protocol 2</b>: Testing for gene essentiality using CRISPRi spot assays on plates</p><p><b>Support Protocol 3</b>: Transformation of <i>E. coli</i> by electroporation</p><p><b>Support Protocol 4</b>: Transformation of CaCl<sub>2</sub>-competent <i>E. coli</i></p>\",\"PeriodicalId\":39967,\"journal\":{\"name\":\"Current Protocols in Microbiology\",\"volume\":\"59 1\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2020-12-17\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://sci-hub-pdf.com/10.1002/cpmc.130\",\"citationCount\":\"7\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current Protocols in Microbiology\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cpmc.130\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"\",\"JCRName\":\"\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current Protocols in Microbiology","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpmc.130","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 7
引用
批量引用
Programmable Gene Knockdown in Diverse Bacteria Using Mobile-CRISPRi
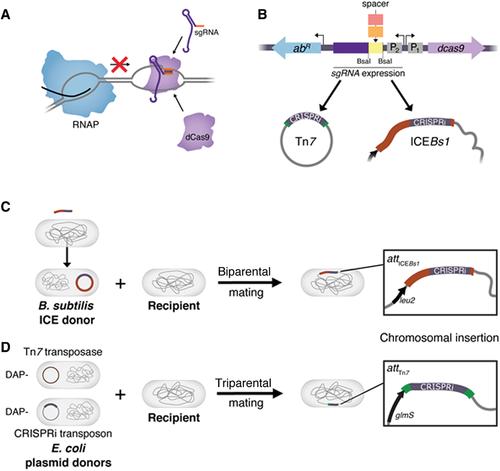
Facile bacterial genome sequencing has unlocked a veritable treasure trove of novel genes awaiting functional exploration. To make the most of this opportunity requires powerful genetic tools that can target all genes in diverse bacteria. CRISPR interference (CRISPRi) is a programmable gene-knockdown tool that uses an RNA-protein complex comprised of a single guide RNA (sgRNA) and a catalytically inactive Cas9 nuclease (dCas9) to sterically block transcription of target genes. We previously developed a suite of modular CRISPRi systems that transfer by conjugation and integrate into the genomes of diverse bacteria, which we call Mobile-CRISPRi. Here, we provide detailed protocols for the modification and transfer of Mobile-CRISPRi vectors for the purpose of knocking down target genes in bacteria of interest. We further discuss strategies for optimizing Mobile-CRISPRi knockdown, transfer, and integration. We cover the following basic protocols: sgRNA design, cloning new sgRNA spacers into Mobile-CRISPRi vectors, Tn7 transfer of Mobile-CRISPRi to Gram-negative bacteria, and ICEBs1 transfer of Mobile-CRISPRi to Bacillales. © 2020 The Authors.
Basic Protocol 1 : sgRNA design
Basic Protocol 2 : Cloning of new sgRNA spacers into Mobile-CRISPRi vectors
Basic Protocol 3 : Tn7 transfer of Mobile-CRISPRi to Gram-negative bacteria
Basic Protocol 4 : ICEBs1 transfer of Mobile-CRISPRi to Bacillales
Support Protocol 1 : Quantification of CRISPRi repression using fluorescent reporters
Support Protocol 2 : Testing for gene essentiality using CRISPRi spot assays on plates
Support Protocol 3 : Transformation of E. coli by electroporation
Support Protocol 4 : Transformation of CaCl2 -competent E. coli