Overeating and inactivity are associated with type 2 diabetes. This study aimed to investigate its pathological basis using integrated omics and db/db/mice, a model representing this condition.
The study involved housing 8-week-old db/m and db/db mice for 8 weeks. Various analyses were conducted, including gene expression in skeletal muscle and small intestine using next-generation sequencing; cytokine arrays of serum; assessment of metabolites in skeletal muscle, stool, and serum; and analysis of the gut microbiota. Histone modifications in small intestinal epithelial cells were profiled using CUT&Tag.
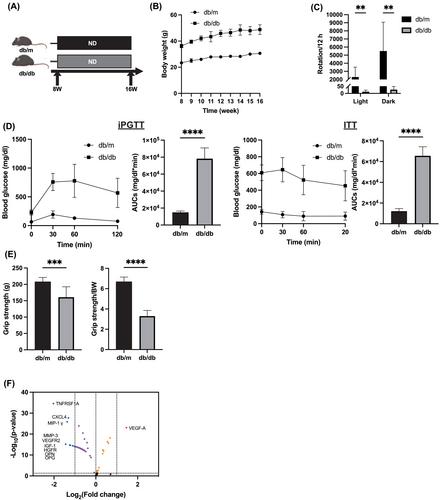
Compared with db/m mice, db/db mice had 22.4% lower grip strength and approximately five times the visceral fat weight (P < 0.0001). Serum cytokine arrays showed a 2.8-fold relative concentration of VEGF-A in db/db mice (P < 0.0001) and lower concentrations of several other cytokines. mRNA sequencing revealed downregulation of Myh expression in skeletal muscle, upregulation of lipid and glucose transporters, and downregulation of amino acid transporters in the small intestine of db/db/mice. The concentrations of saturated fatty acids in skeletal muscle were significantly higher, and the levels of essential amino acids were lower in db/db mice. Analysis of the gut microbiota, 16S rRNA sequencing, revealed lower levels of the phylum Bacteroidetes (59.7% vs. 44.9%) and higher levels of the phylum Firmicutes (20.9% vs. 31.4%) in db/db mice (P = 0.003). The integrated signal of histone modifications of lipid and glucose transporters was higher, while the integrated signal of histone modifications of amino acid transporters was lower in the db/db mice.
The multi-omics approach provided insights into the epigenomic alterations in the small intestine, suggesting their involvement in the pathogenesis of inactivity-induced muscle atrophy in obese mice.