Sarcopenia is an age-related muscle disease that increases the risk of falls, disabilities, and death. It is associated with increased muscle protein degradation driven by molecular signalling pathways including Akt and FOXO1. This study aims to identify genes, gene interactions, and molecular pathways and processes associated with muscle aging and exercise in older adults that remained undiscovered until now leveraging on an artificial intelligence approach called artificial neural network inference (ANNi).
Four datasets reporting the profile of muscle transcriptome obtained by RNA-seq of young (21–43 years) and older adults (63–79 years) were selected and retrieved from the Gene Expression Omnibus (GEO) data repository. Two datasets contained the transcriptome profiles associated to muscle aging and two the transcriptome linked to resistant exercise in older adults, the latter before and after 6 months of exercise training. Each dataset was individually analysed by ANNi based on a swarm neural network approach integrated into a deep learning model (Intelligent Omics). This allowed us to identify top 200 genes influencing (drivers) or being influenced (targets) by aging or exercise and the strongest interactions between such genes. Downstream gene ontology (GO) analysis of these 200 genes was performed using Metacore (Clarivate™) and the open-source software, Metascape. To confirm the differential expression of the genes showing the strongest interactions, real-time quantitative PCR (RT-qPCR) was employed on human muscle biopsies obtained from eight young (25 ± 4 years) and eight older men (78 ± 7.6 years), partaking in a 6-month resistance exercise training programme.
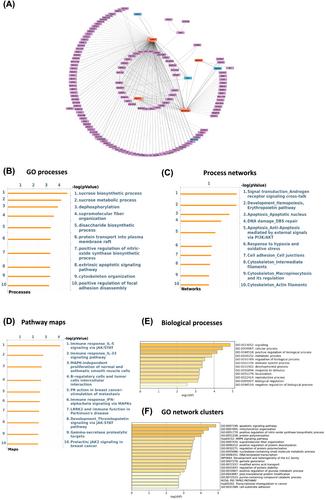
CHAD, ZDBF2, USP54, and JAK2 were identified as the genes with the strongest interactions predicting aging, while SCFD1, KDM5D, EIF4A2, and NIPAL3 were the main interacting genes associated with long-term exercise in older adults. RT-qPCR confirmed significant upregulation of USP54 (P = 0.005), CHAD (P = 0.03), and ZDBF2 (P = 0.008) in the aging muscle, while exercise-related genes were not differentially expressed (EIF4A2 P = 0.99, NIPAL3 P = 0.94, SCFD1 P = 0.94, and KDM5D P = 0.64). GO analysis related to skeletal muscle aging suggests enrichment of pathways linked to bone development (adj P-value 0.006), immune response (adj P-value <0.001), and apoptosis (adj P-value 0.01). In older exercising adults, these were ECM remodelling (adj P-value <0.001), protein folding (adj P-value <0.001), and proteolysis (adj P-value <0.001).
Using ANNi and RT-qPCR, we identified three strongly interacting genes predicting muscle aging, ZDBF2, USP54, and CHAD. These findings can help to inform the design of nonpharmacological and pharmacological interventions that prevent or mitigate sarcopenia.