Targeted Protein Degradation Phenotypic Studies Using HaloTag CRISPR/Cas9 Endogenous Tagging Coupled with HaloPROTAC3
Elizabeth A. Caine, Sarah D. Mahan, Rebecca L. Johnson, Amanda N. Nieman, Ngan Lam, Curtis R. Warren, Kristin M. Riching, Marjeta Urh, Danette L. Daniels
下载PDF
{"title":"Targeted Protein Degradation Phenotypic Studies Using HaloTag CRISPR/Cas9 Endogenous Tagging Coupled with HaloPROTAC3","authors":"Elizabeth A. Caine, Sarah D. Mahan, Rebecca L. Johnson, Amanda N. Nieman, Ngan Lam, Curtis R. Warren, Kristin M. Riching, Marjeta Urh, Danette L. Daniels","doi":"10.1002/cpph.81","DOIUrl":null,"url":null,"abstract":"<p>To assess the role of a protein, protein loss phenotypic studies can be used, most commonly through mutagenesis RNAi or CRISPR knockout. Such studies have been critical for the understanding of protein function and the identification of putative therapeutic targets for numerous human disease states. However, these methodological approaches present challenges because they are not easily reversible, and if an essential gene is targeted, an associated loss of cell viability can potentially hinder further studies. Here we present a reversible and conditional live-cell knockout strategy that is applicable to numerous proteins. This modular protein-tagging approach regulates target loss at the protein, rather than the genomic, level through the use of HaloPROTAC3, which specifically degrades HaloTag fusion proteins via recruitment of the VHL E3 ligase component. To enable HaloTag-mediated degradation of endogenous proteins, we provide protocols for HaloTag genomic insertion at the protein N or C terminus via CRISPR/Cas9 and use of HaloTag fluorescent ligands to enrich edited cells via Fluorescence-Activated Cell Sorting (FACS). Using these approaches, endogenous HaloTag fusion proteins present in various subcellular locations can be degraded by HaloPROTAC3. As detecting the degradation of endogenous targets is challenging, the 11-amino-acid peptide tag HiBiT is added to the HaloTag fusion to allows the sensitive luminescence detection of HaloTag fusion levels without the use of antibodies. Lastly, we demonstrate, through comparison of HaloPROTAC3 degradation with that of another fusion tag PROTAC, dTAG-13, that HaloPROTAC3 has a faster degradation rate and similar extent of degradation. © 2020 The Authors.</p><p><b>Basic Protocol 1</b>: CRISPR/Cas9 insertion of HaloTag or HiBiT-HaloTag</p><p><b>Basic Protocol 2</b>: HaloPROTAC3 degradation of endogenous HaloTag fusions</p>","PeriodicalId":10871,"journal":{"name":"Current Protocols in Pharmacology","volume":"91 1","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2020-12-17","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cpph.81","citationCount":"12","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current Protocols in Pharmacology","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpph.81","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q2","JCRName":"Pharmacology, Toxicology and Pharmaceutics","Score":null,"Total":0}
引用次数: 12
引用
批量引用
Abstract
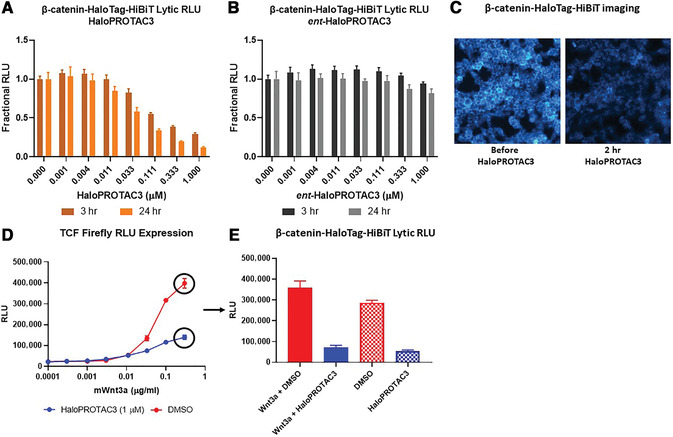
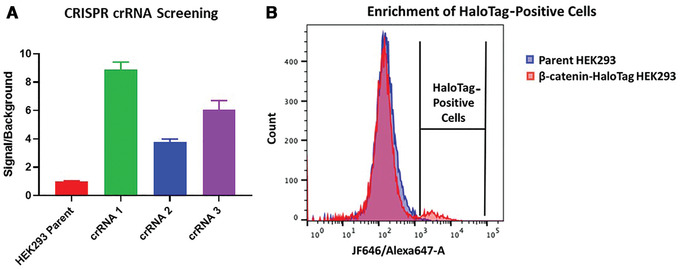
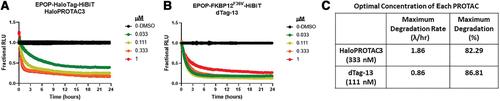
To assess the role of a protein, protein loss phenotypic studies can be used, most commonly through mutagenesis RNAi or CRISPR knockout. Such studies have been critical for the understanding of protein function and the identification of putative therapeutic targets for numerous human disease states. However, these methodological approaches present challenges because they are not easily reversible, and if an essential gene is targeted, an associated loss of cell viability can potentially hinder further studies. Here we present a reversible and conditional live-cell knockout strategy that is applicable to numerous proteins. This modular protein-tagging approach regulates target loss at the protein, rather than the genomic, level through the use of HaloPROTAC3, which specifically degrades HaloTag fusion proteins via recruitment of the VHL E3 ligase component. To enable HaloTag-mediated degradation of endogenous proteins, we provide protocols for HaloTag genomic insertion at the protein N or C terminus via CRISPR/Cas9 and use of HaloTag fluorescent ligands to enrich edited cells via Fluorescence-Activated Cell Sorting (FACS). Using these approaches, endogenous HaloTag fusion proteins present in various subcellular locations can be degraded by HaloPROTAC3. As detecting the degradation of endogenous targets is challenging, the 11-amino-acid peptide tag HiBiT is added to the HaloTag fusion to allows the sensitive luminescence detection of HaloTag fusion levels without the use of antibodies. Lastly, we demonstrate, through comparison of HaloPROTAC3 degradation with that of another fusion tag PROTAC, dTAG-13, that HaloPROTAC3 has a faster degradation rate and similar extent of degradation. © 2020 The Authors.
Basic Protocol 1 : CRISPR/Cas9 insertion of HaloTag or HiBiT-HaloTag
Basic Protocol 2 : HaloPROTAC3 degradation of endogenous HaloTag fusions
利用HaloTag CRISPR/Cas9内源性标记与HaloPROTAC3偶联的靶向蛋白降解表型研究
为了评估蛋白质的作用,可以使用蛋白质损失表型研究,最常见的是通过诱变RNAi或CRISPR敲除。这些研究对于了解蛋白质功能和确定许多人类疾病状态的假定治疗靶点至关重要。然而,这些方法学方法存在挑战,因为它们不容易逆转,如果一个基本基因被靶向,细胞活力的相关丧失可能会阻碍进一步的研究。在这里,我们提出了一种可逆和有条件的活细胞敲除策略,适用于许多蛋白质。这种模块化的蛋白质标记方法通过使用HaloPROTAC3在蛋白质而不是基因组水平上调节靶标丢失,HaloPROTAC3通过募集VHL E3连接酶组分特异性地降解HaloTag融合蛋白。为了实现HaloTag介导的内源性蛋白降解,我们提供了通过CRISPR/Cas9将HaloTag基因组插入蛋白N或C端,并使用HaloTag荧光配体通过荧光活化细胞分选(FACS)富集编辑细胞的方案。利用这些方法,存在于不同亚细胞位置的内源性HaloTag融合蛋白可以被HaloPROTAC3降解。由于检测内源性靶标的降解具有挑战性,因此在HaloTag融合中添加了11-氨基酸肽标签HiBiT,以便在不使用抗体的情况下对HaloTag融合水平进行敏感的发光检测。最后,通过与另一种融合标签PROTAC dTAG-13的降解比较,我们证明HaloPROTAC3具有更快的降解速度和相似的降解程度。©2020作者。基本方案1:CRISPR/Cas9插入HaloTag或hibit - halotag2:内源性HaloTag融合物降解HaloPROTAC3
本文章由计算机程序翻译,如有差异,请以英文原文为准。