Genome-wide association studies (GWAS) have identified hundreds of common variants associated with alcohol consumption. In contrast, genetic studies of alcohol consumption that use rare variants are still in their early stages. No prior studies of alcohol consumption have examined whether common and rare variants implicate the same genes and molecular networks, leaving open the possibility that the two approaches might identify distinct biology.
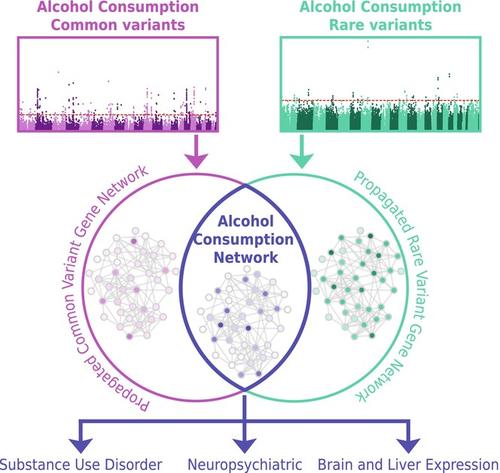
To address this knowledge gap, we used publicly available alcohol consumption GWAS summary statistics (GSCAN, N = 666,978) and whole exome sequencing data (Genebass, N = 393,099) to identify a set of common and rare variants for alcohol consumption. We used gene-based analysis to implicate genes from common and rare variant analyses, which we then propagated onto a shared molecular network using a network colocalization procedure.
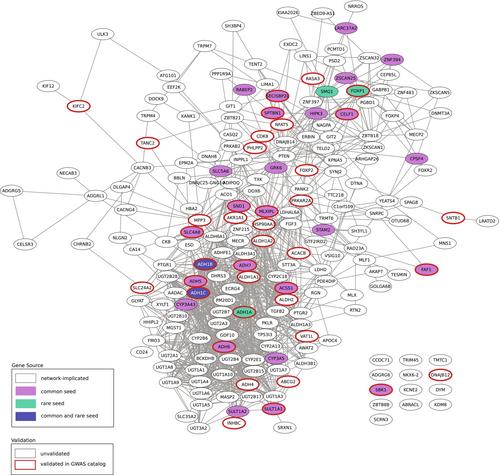
Gene-based analysis of each dataset implicated 294 (common variants) and 35 (rare variants) genes, including ethanol metabolizing genes ADH1B and ADH1C, which were identified by both analyses, and ANKRD12, GIGYF1, KIF21B, and STK31, which were identified in only the rare variant analysis, but have been associated with other neuropsychiatric traits. Network colocalization revealed significant network overlap between the genes identified via common and rare variants. The shared network identified gene families that function in alcohol metabolism, including ADH, ALDH, CYP, and UGT. Seventy-one of the genes in the shared network were previously implicated in neuropsychiatric or substance use disorders but not alcohol-related behaviors (e.g. EXOC2, EPM2A, and CACNG4). Differential gene expression analysis showed enrichment in the liver and several brain regions.
Genes implicated by network colocalization identify shared biology relevant to alcohol consumption, which also underlie neuropsychiatric traits and substance use disorders that are comorbid with alcohol use, providing a more holistic understanding of two disparate sources of genetic information.